Bayesian Models with Multiple Predictors

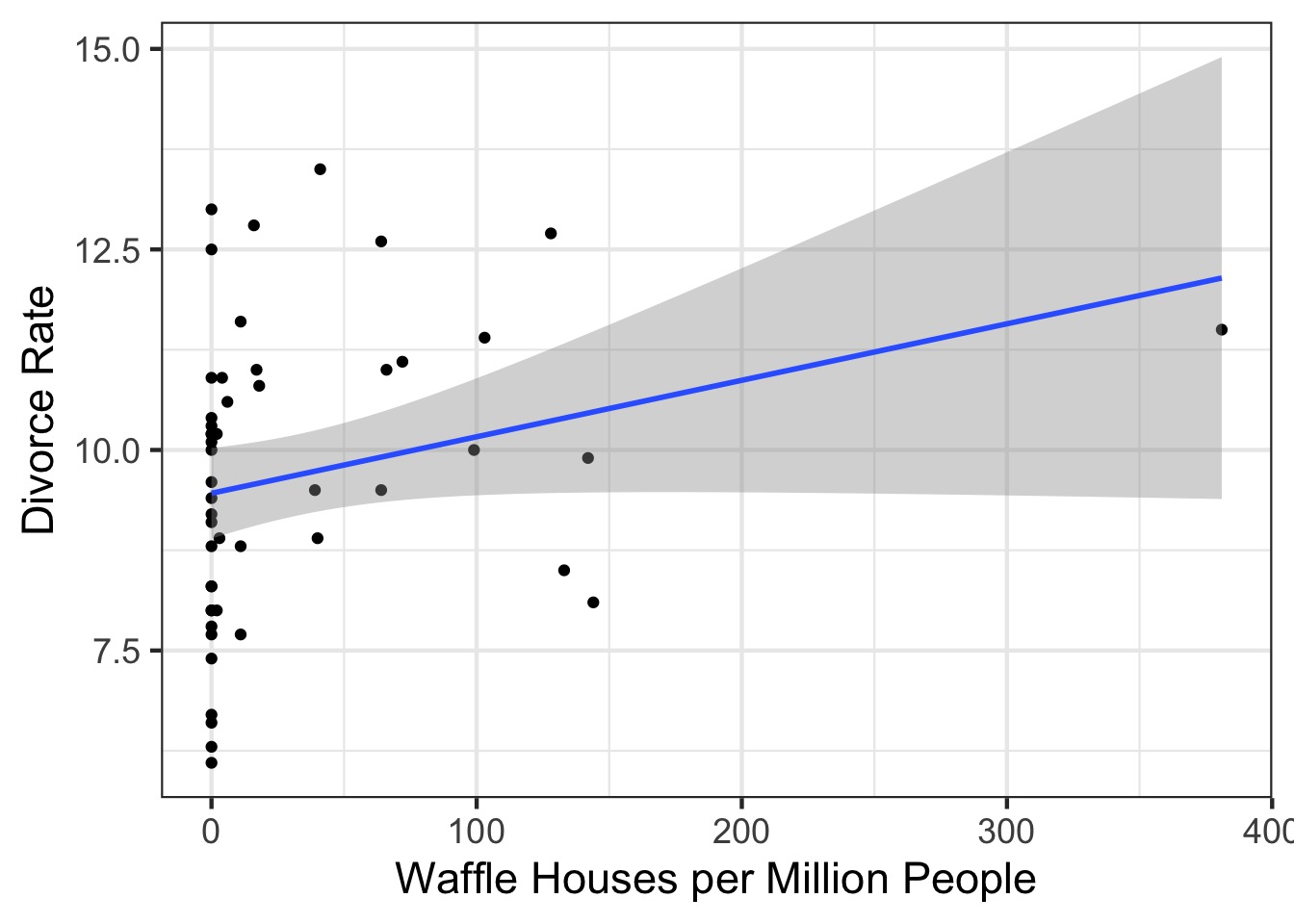
Waffle House: Does it Lead to Perdition?
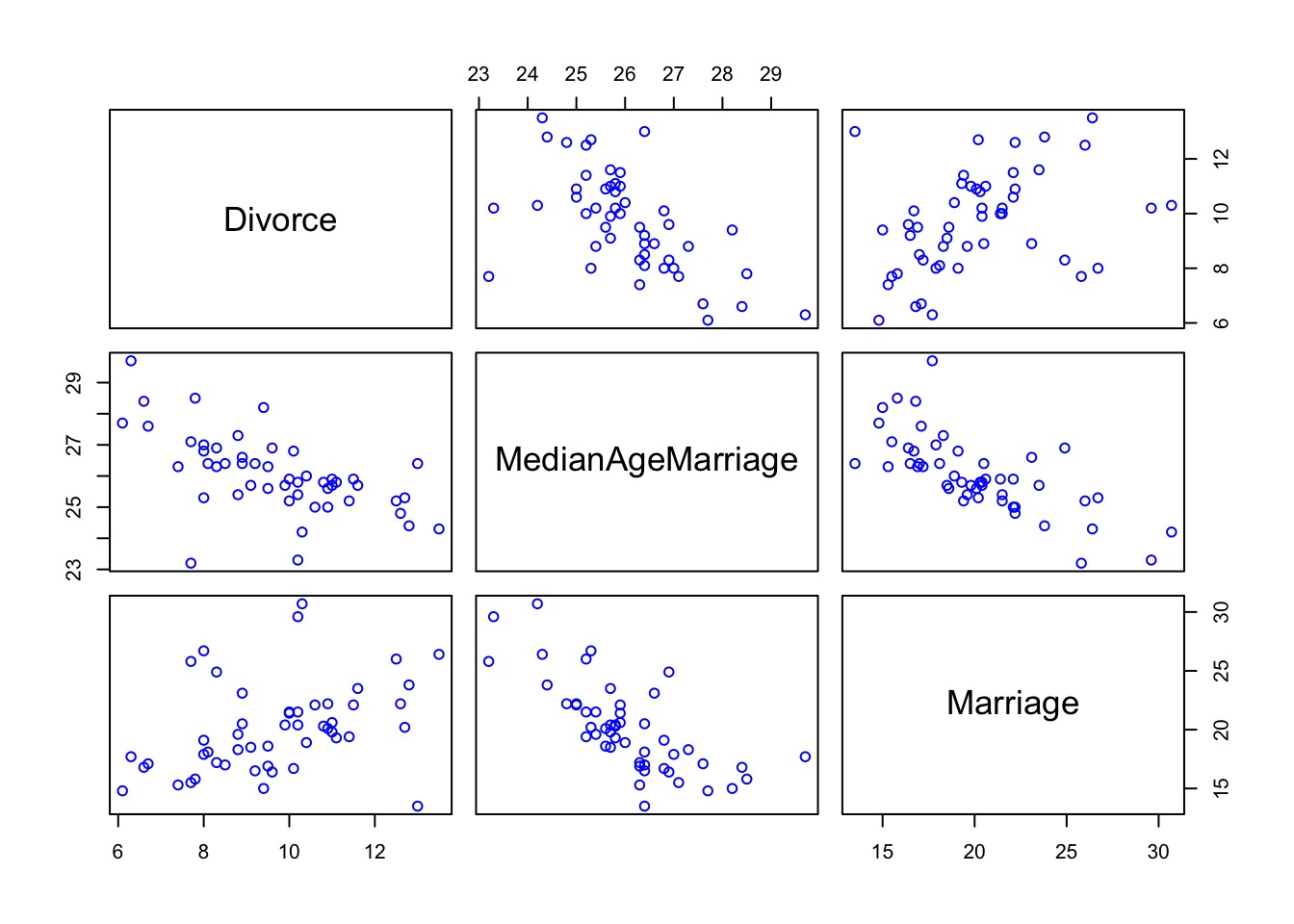
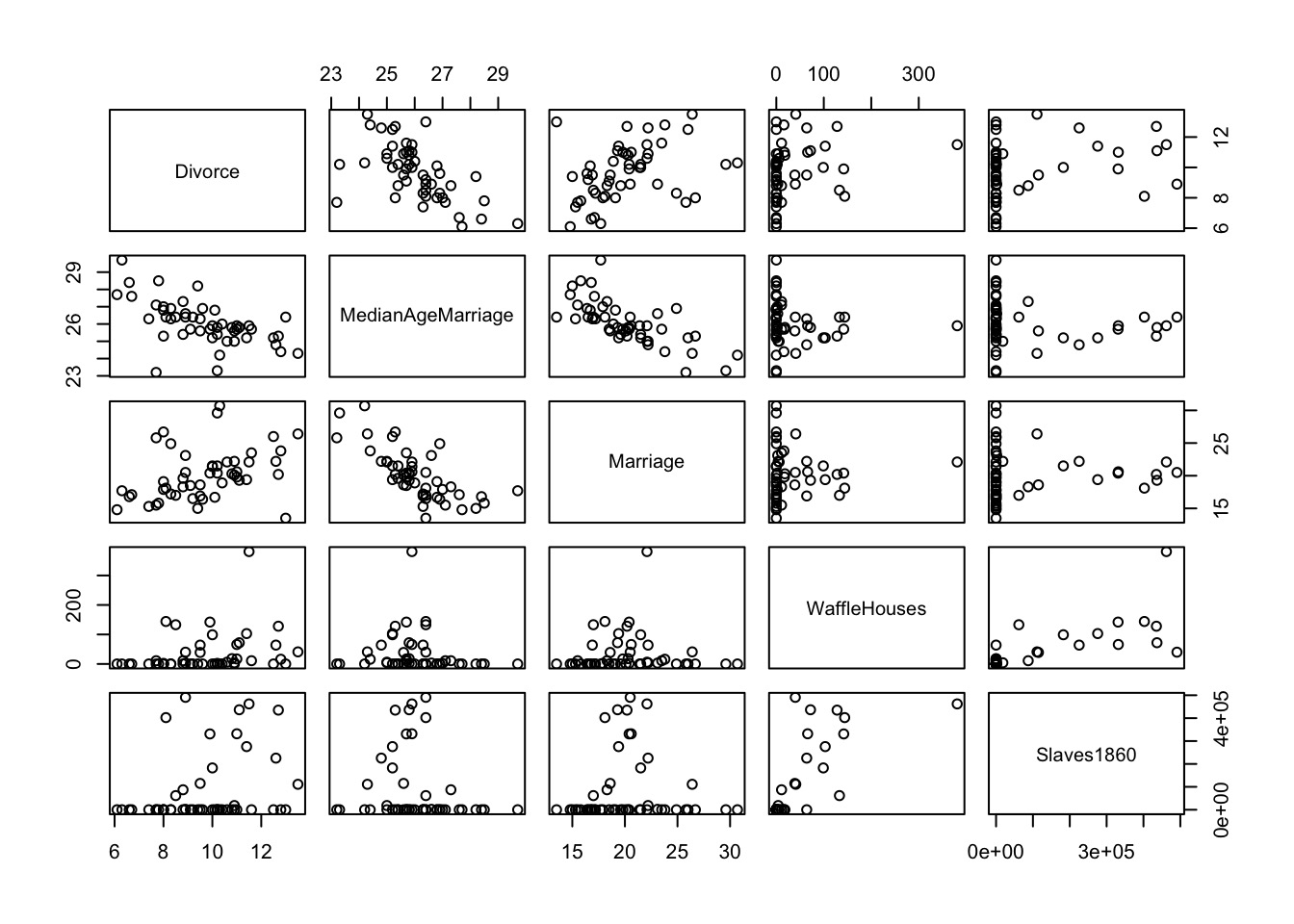
So many possibilities
Today’s Outline
Multiple Predictors in a Bayesian Framework
- How multiple predictors tease apart spurious and masked relationships
Evaluating a Model with Multiple Predictors
Testing Mediation
Categorical Variables
Why use multiple predictors
Controlling for confounds
- Disentangle spurious relationships
- Reveal masked relationships
Dealing with multiple causation
Interactions (soon)
Why NOT use multiple predictors
Multicollinearity
- But can aggregate variables
Overfitting
- But see model selection
Loss of precision in estimates
Interpretability

DAG Before You Model
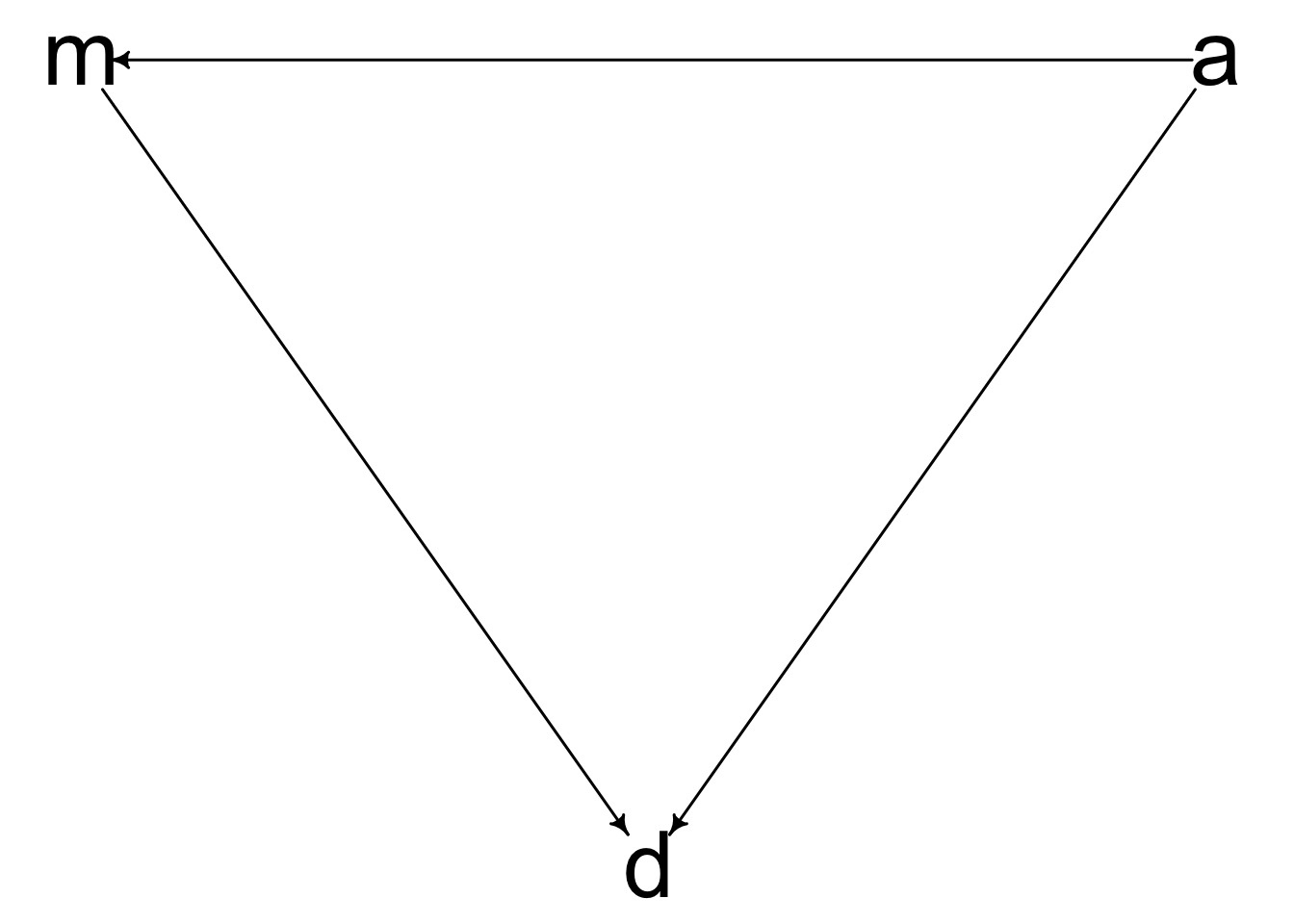
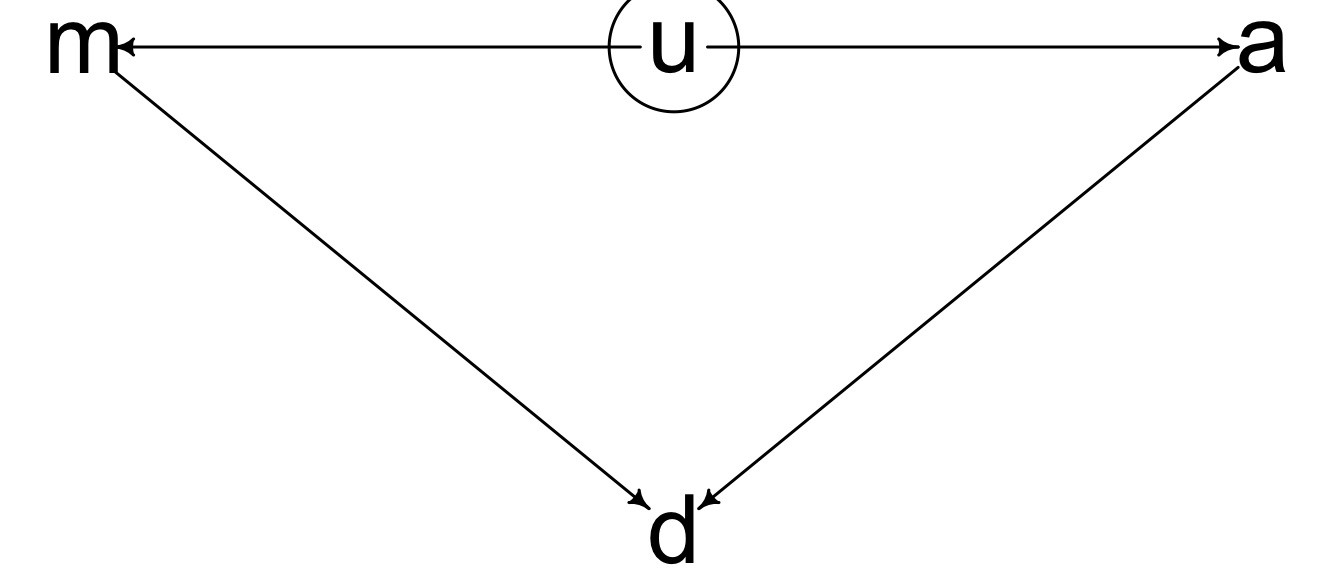
Which of these is your model?

Should we Standardize?
- Standardization can aid in fitting.
- No scaling issues
- Faster convergence
- Reduces collinearity of derived predictors (nonlinear terms)
- No scaling issues
- Standardization X and/or Y can aid in choosing priors
- Centering moves intercept to 0 on X and/or Y axis
- Dividing by SD means we can think in terms of standard normal for slopes and intercepts
- Centering moves intercept to 0 on X and/or Y axis
- Interpretation
- For linear models, coefficients are standardized correlation coefficients
- Can back-transform Y by multiplying by sd and adding mean
- For linear models, coefficients are standardized correlation coefficients
Let’s Standardize Divorce
Or use standardize() or write a function
How to Build a Model with Multiple Predictors
Likelihood:
\(y_i \sim Normal(\mu_i, \sigma)\)
Data Generating Process
\(\mu_i = \alpha + \beta_1 x1_i + \beta_2 x2_i + ...\)
Prior:
\(\alpha \sim Normal(0, 0.5)\)
\(\beta_j \sim Normal(0, 0.5)\) prior for each
\(\sigma \sim Exp(1)\)
Our Model
Likelihood:
\(D_i \sim Normal(\mu_i, \sigma)\)
Data Generating Process
\(\mu_i = \alpha + \beta_m M_i + \beta_a A_i\)
Prior:
\(\alpha \sim Normal(0, 0.5)\) because standardized
\(\beta_m \sim Normal(0, 0.5)\) because standardized
\(\beta_a \sim Normal(0, 0.5)\) because standardized
\(\sigma \sim Exp(1)\) because standardized
Our Model
Were Our Priors Reasonable?
Prior for Marriage Age N(0,0.5)
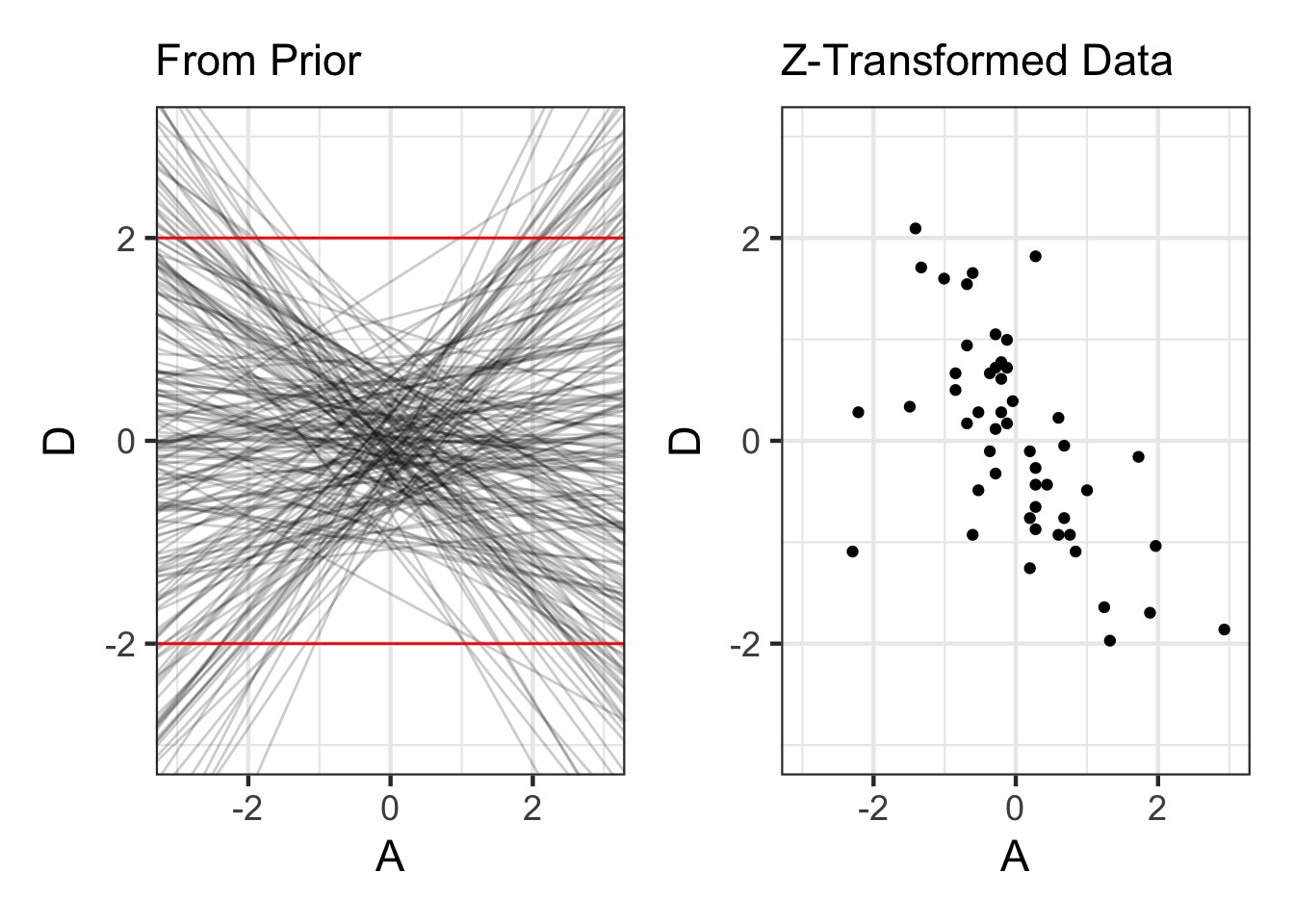
Prior and Plot
prior_divorce <- extract.prior(fit, n = 200) |>
as_tibble()
p1 <- ggplot(data = prior_divorce) +
geom_abline(aes(slope = bA, intercept = a), alpha = 0.2) +
xlim(c(-3,3)) + ylim(c(-3,3)) +
geom_hline(yintercept = c(-2,2), color = "red") +
labs(y = "D", x = "A", subtitle = "From Prior")
d1 <- ggplot(data = WaffleDivorce) +
geom_point(aes(x = A, y = D)) +
xlim(c(-3,3)) + ylim(c(-3,3)) +
labs(subtitle = "Z-Transformed Data")
p1 + d1Were Our Priors Reasonable?
Prior for Marriage Rate N(0, 0.5)
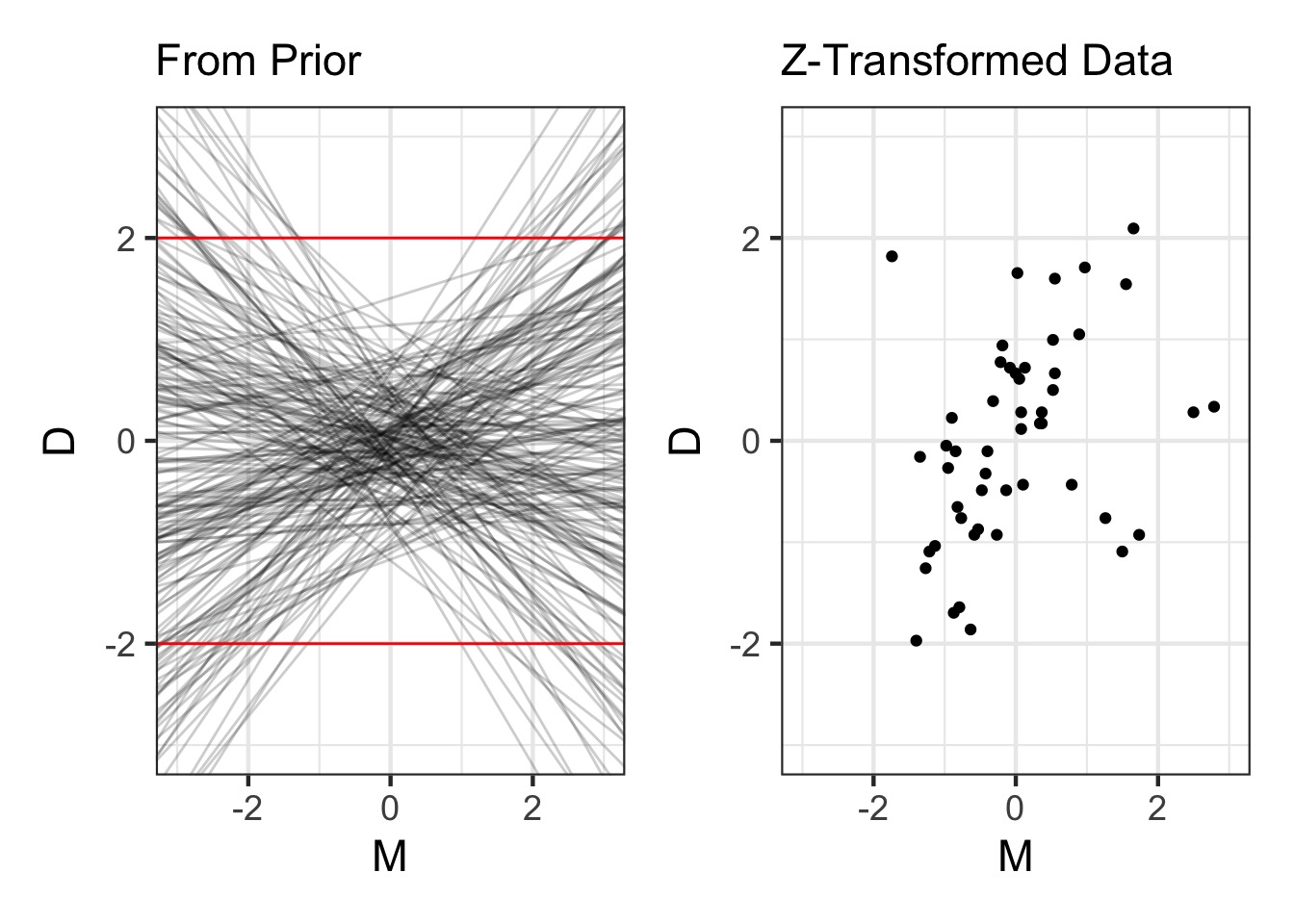
Results: We Only Need Median Age

What does a Multiple Regression Coefficient Mean?
What is the predictive value of one variable once all others have been accounted for?
We want a coefficient that explains the unique contribution of a predictor
What is the effect of x1 on y after we take out the effect of x2 on x1?
Today’s Outline
Multiple Predictors in a Bayesian Framework
- How multiple predictors tease apart spurious and masked relationships
Evaluating a Model with Multiple Predictors
Testing Mediation
Categorical Variables
How to Understand Posteriors
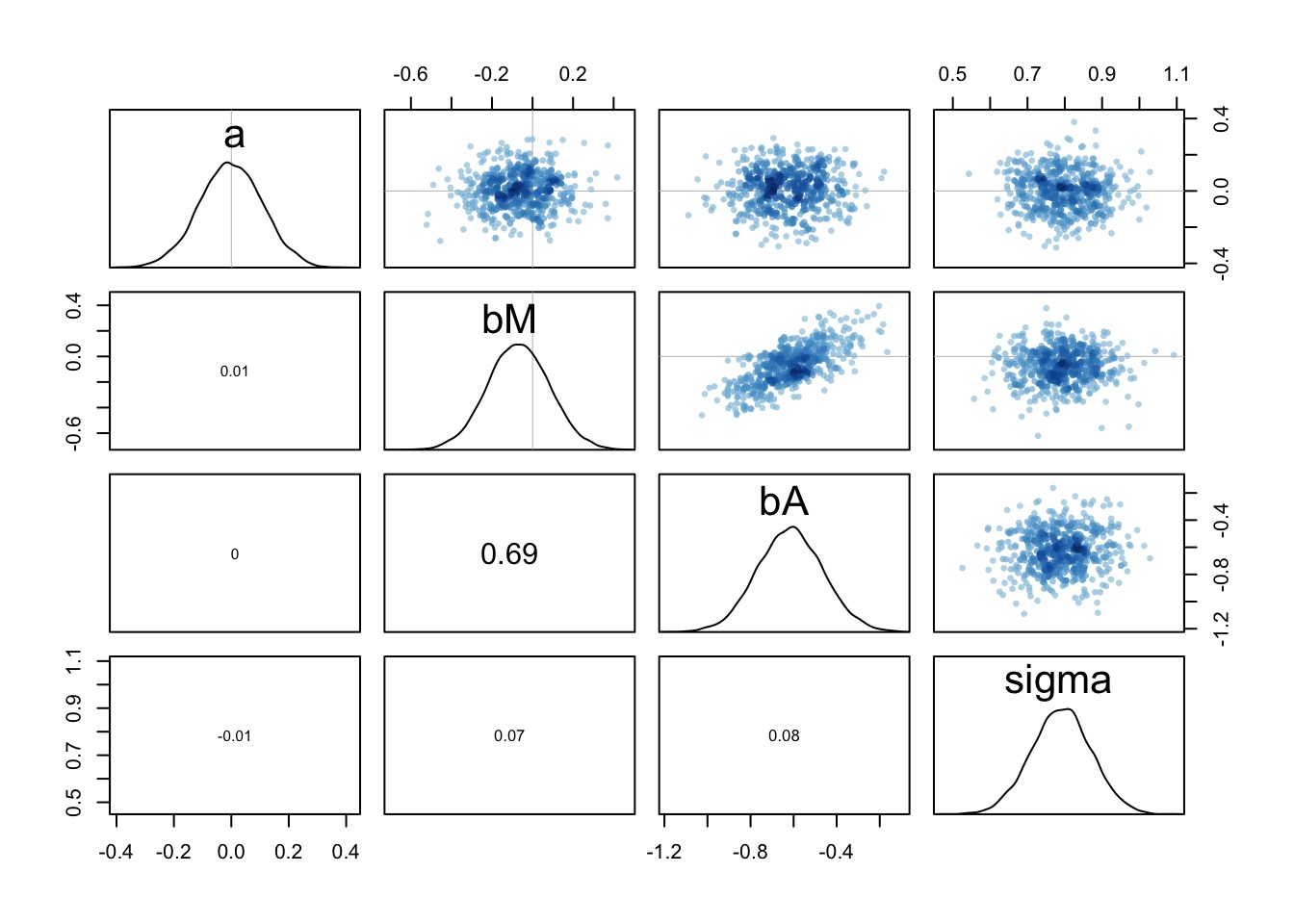
Note correlation between bM and bA - when one is high the other is as well, but see scale
How to Understand Posteriors
- Predictor-residual plots
- What if you remove the effect of other predictors?
- Counterfactual plots
- What if something else had happened?
- Posterior Predictions
- How close are model predictions to the data
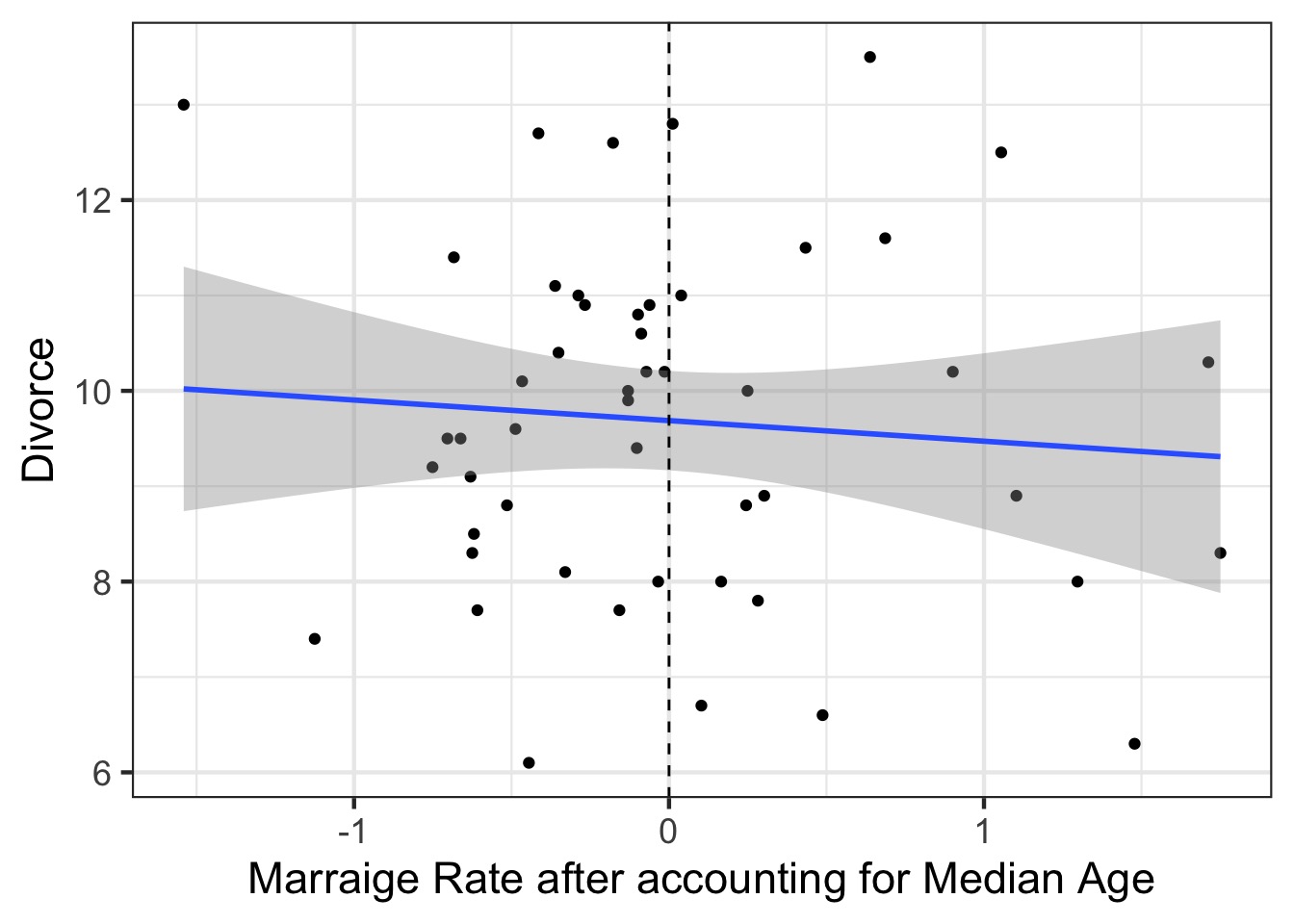
Predictor-Residual Plots
- The
cr.plotsfrom thecarpackage- Component-residual
- Take the residual of a predictor, assess it’s predictive power
Steps to Make Predictor-Residual Plots
Compute predictor 1 ~ all other predictors
Take residual of predictor 1
Regress predictor 1 on response
PR Model Part 1
PR Model Part 2
The Predictor-Residual Plot
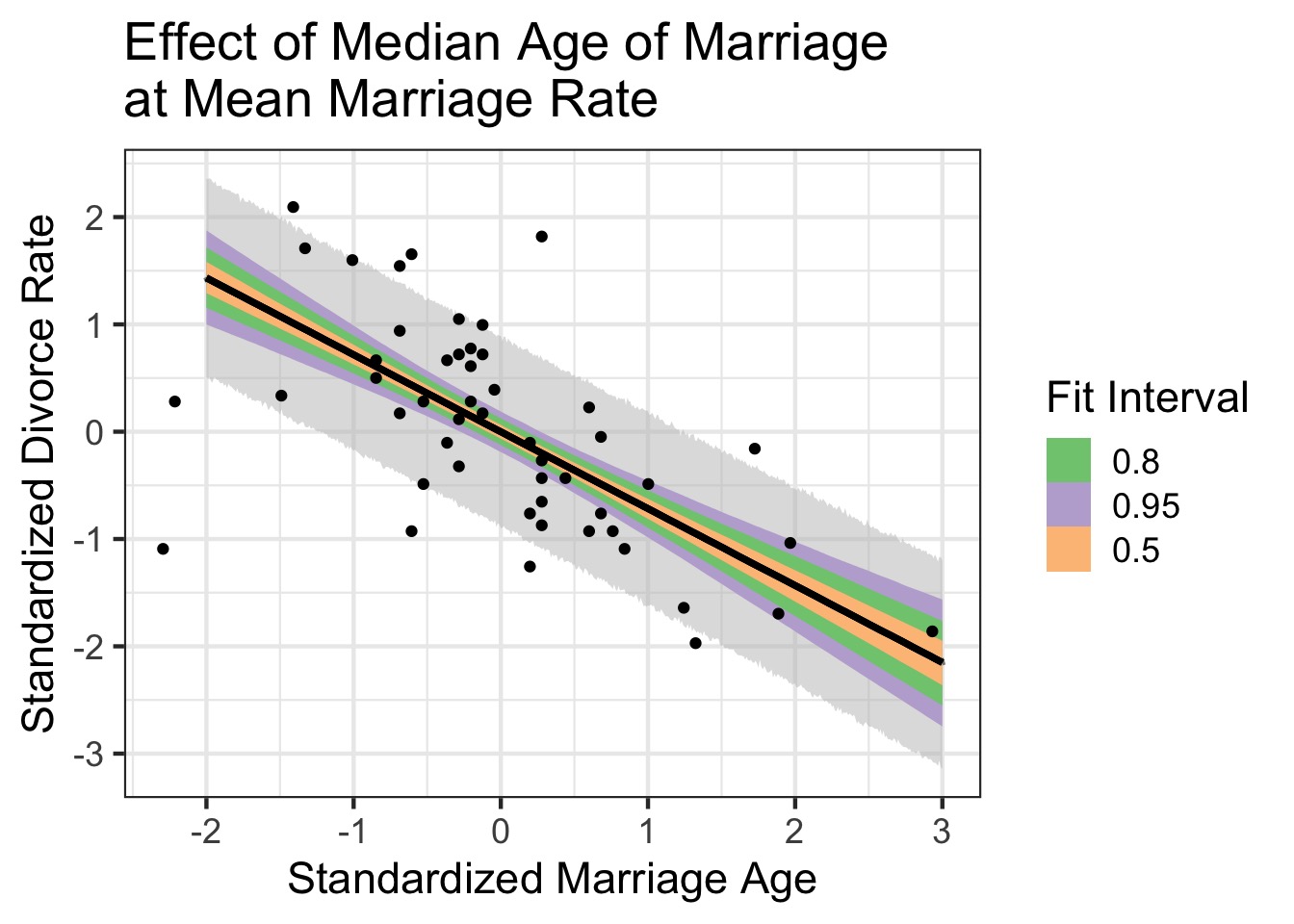
Counterfactual Plots
Counterfactual: A conditional statement of “if this, then …”
Powerful way of assessing models - “If we had seen Marriage Rate as x, then the effect of Median age on divorce rate would be…”
Shows model implied predictions, often at levels nor observed
But - WHICH if?
Estimate effect of Age and Rate controlling for one another

Estimate total, direct, & indirect effect of Age on Divorce

(This is Structural Equation Modeling)
Counterfactual Plots: Code
Effect of Age on Divorce holding Rate at it’s Mean
What do we learn about the effects of Median Marriage Age Alone?
Today’s Outline
Multiple Predictors in a Bayesian Framework
- How multiple predictors tease apart spurious and masked relationships
Evaluating a Model with Multiple Predictors
Testing Mediation
Categorical Variables
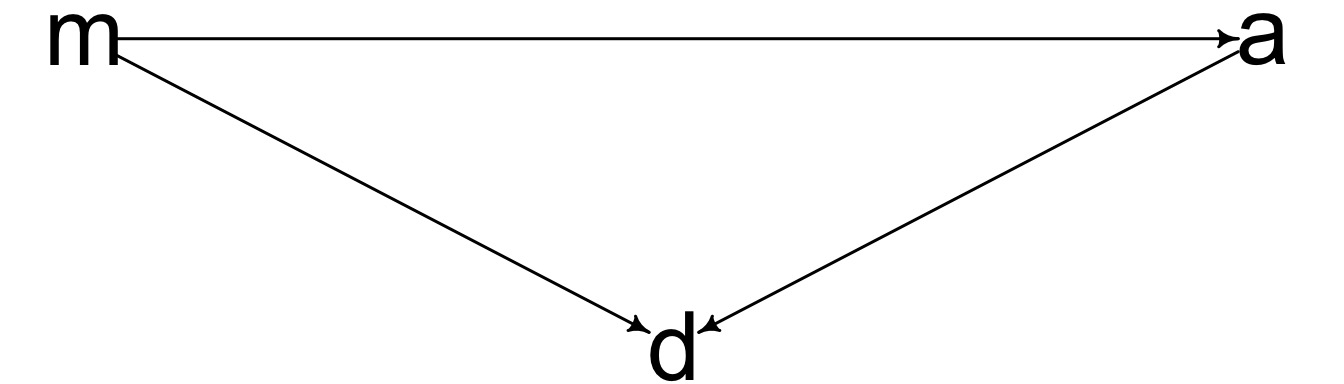
What if this was right?
Mediation
The effect of one variable has a direct and indirect effect
The indirect effect is mediated through another variable.
Multiple types of mediation
- Full mediation (i.e., no direct effect)
- Partial mediation (i.e., a direct and indirect effect)
- Unmediated relationship (direct effect only)
- Full mediation (i.e., no direct effect)
We can look at strength of indirect and direct effects to differentiate
- Or we can use model selection
- Adding indirect effects also allows tests of causal independence
- Or we can use model selection
Our New Model
Likelihoods:
\(D_i \sim Normal(\mu_{di}, \sigma_d)\)
\(M_i \sim Normal(\mu_{mi}, \sigma_m)\)
Data Generating Processes
\(\mu_{di} = \alpha_d + \beta_m M_i + \beta_a A_i\)
\(\mu_{mi} = \alpha_m + \beta_{ma} A_i\)
Prior:
\(\alpha_d \sim Normal(0, 0.5)\)
\(\alpha_m \sim Normal(0, 0.5)\)
\(\beta_m \sim Normal(0, 0.5)\)
\(\beta_a \sim Normal(0, 0.5)\)
\(\beta_{ma} \sim Normal(0, 0.5)\)
\(\sigma_d \sim Exp(1)\)
\(\sigma_m \sim Exp(1)\)
Our Model
mod_med <- alist(
## A -> D <- M
#likelihood
D ~ dnorm(mu, sigma),
#data generating processes
mu <- a + bM*M + bA * A,
# Priors
a ~ dnorm(0, 0.5),
bM ~ dnorm(0, 0.5),
bA ~ dnorm(0, 0.5),
sigma ~ dunif(0,10),
## A -> M
#likelihood
M ~ dnorm(mu_m, sigma_m),
#data generating processes
mu_m <- a_m + bMA*A,
# Priors
a_m ~ dnorm(0, 0.5),
bMA ~ dnorm(0, 0.5),
sigma_m ~ dunif(0,10)
)
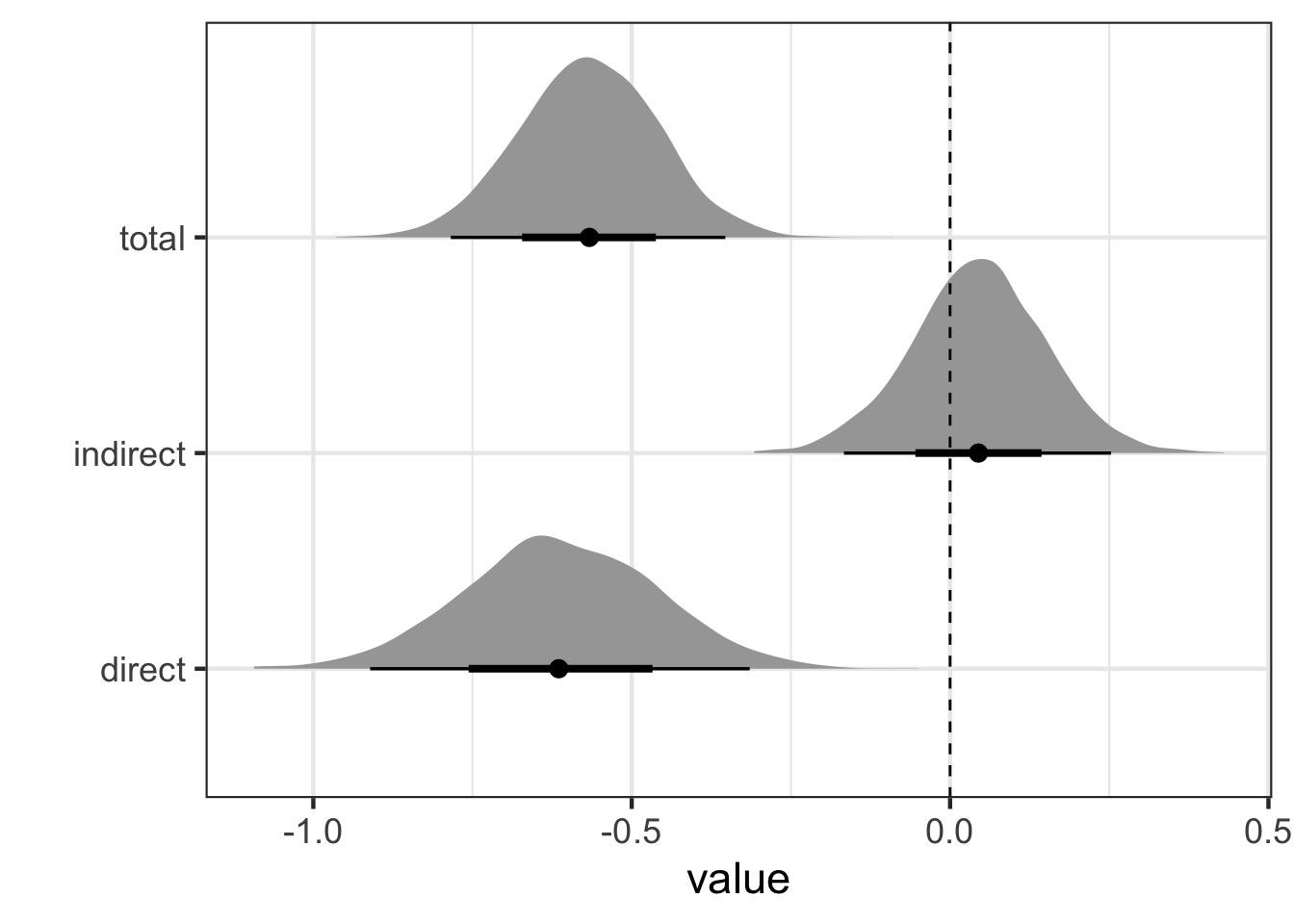
fit_med <- quap(mod_med, data=WaffleDivorce)Calculating Direct and Indirect Effects
- Direct: bA
- Indirect: bMA x bM
- Total = bA + bMA x bM
Calculate and Visualize
We can draw coefficients and calculate using our DAG
Is there Mediation here?
Code
Today’s Outline
Multiple Predictors in a Bayesian Framework
Evaluating a Model with Multiple Predictors
Testing Mediation
Categorical Variables
Categorical Variables
Lots of ways to write models with categorical variables
We all hate R’s treatment contrasts
Two main ways to write a model
Categorical Model Construction
- Code each level as 1 or 0 if present/absent
- Need to have one baseline level
- Treatment contrasts!
Y <- a + b * x_is_level
- Need to have one baseline level
- Index your categories
- Need to convert factors to levels with
as.numeric()
y <- a[level]
- Need to convert factors to levels with
Monkies and Milk

Monkies and Milk Production
clade species kcal.per.g perc.fat perc.protein
1 Strepsirrhine Eulemur fulvus 0.49 16.60 15.42
2 Strepsirrhine E macaco 0.51 19.27 16.91
3 Strepsirrhine E mongoz 0.46 14.11 16.85
4 Strepsirrhine E rubriventer 0.48 14.91 13.18
5 Strepsirrhine Lemur catta 0.60 27.28 19.50
6 New World Monkey Alouatta seniculus 0.47 21.22 23.58
perc.lactose mass neocortex.perc
1 67.98 1.95 55.16
2 63.82 2.09 NA
3 69.04 2.51 NA
4 71.91 1.62 NA
5 53.22 2.19 NA
6 55.20 5.25 64.54To easily make Variables
Original Milk Model
Milk Coefs: What does a and b mean?
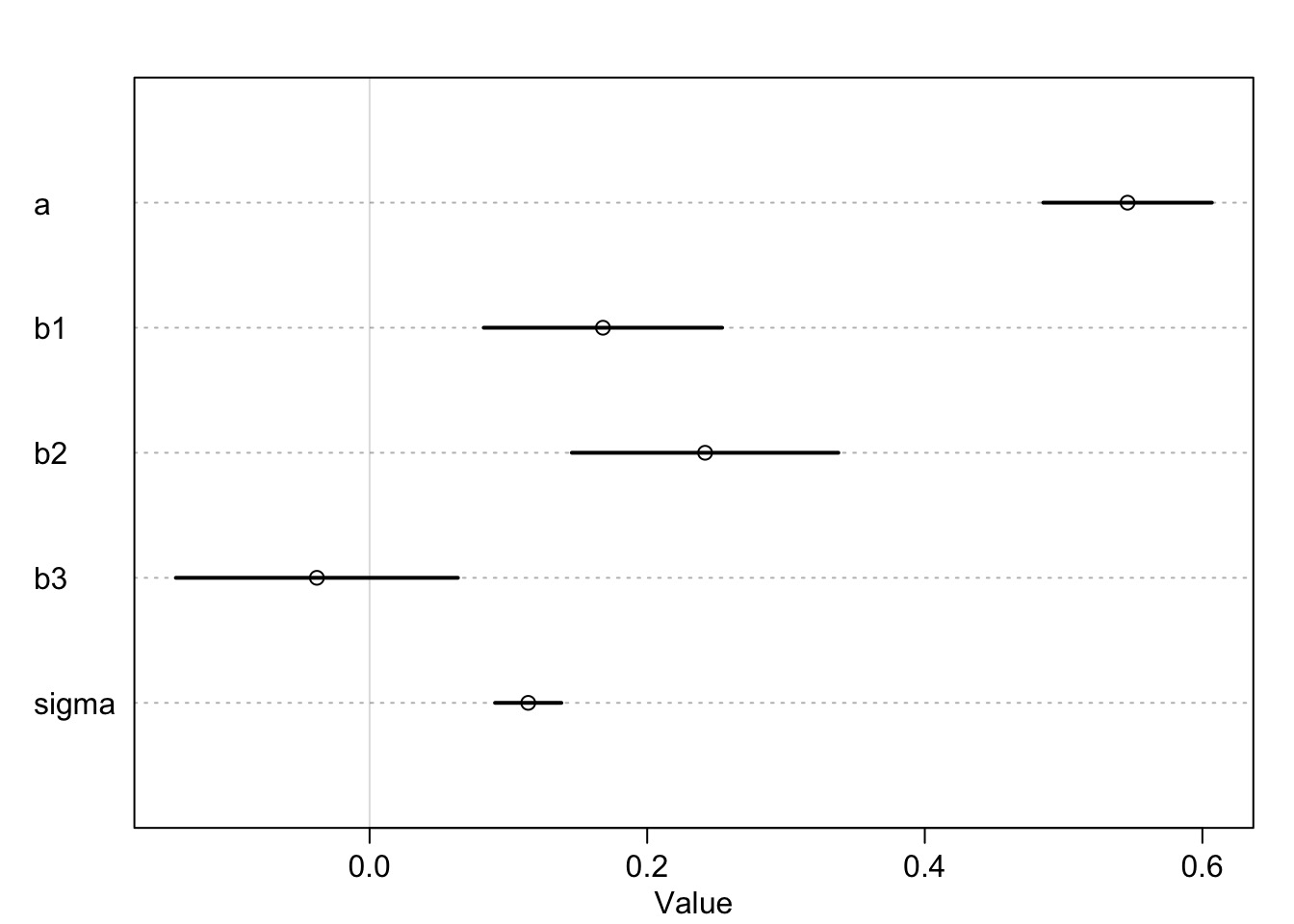
To get the New World mean…
A Better Way
Compare Results
Exercise
Build a model explaing the
kcal.per.gof milkFirst try 2 continuous predictors
Add clade
Bonus: Can you make an interaction (try this last)