Bayesian Linear Regression

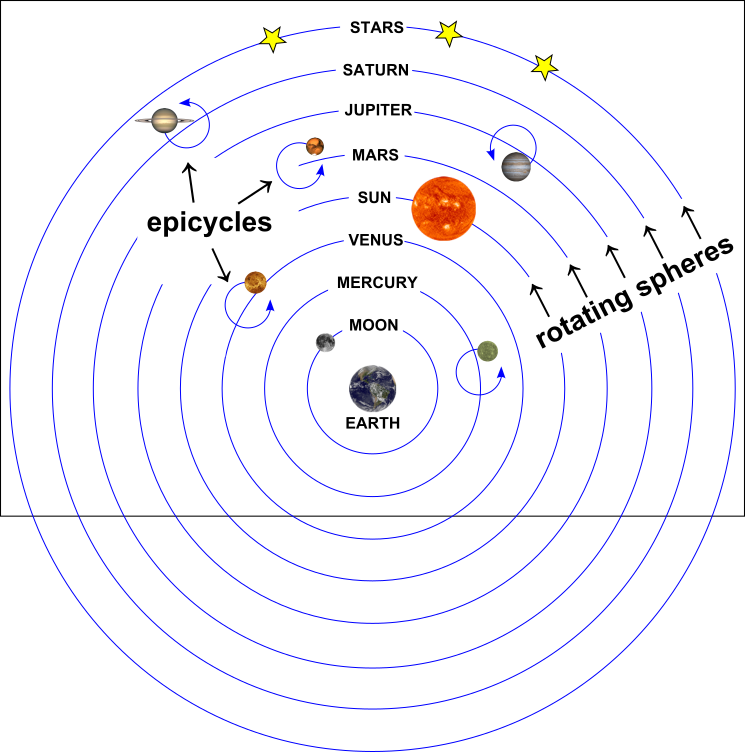
A Ptolemeic Model of the Universe
Why a Normal Error Distribution
- Good descriptor of sum of many small errors
- True for many different distributions
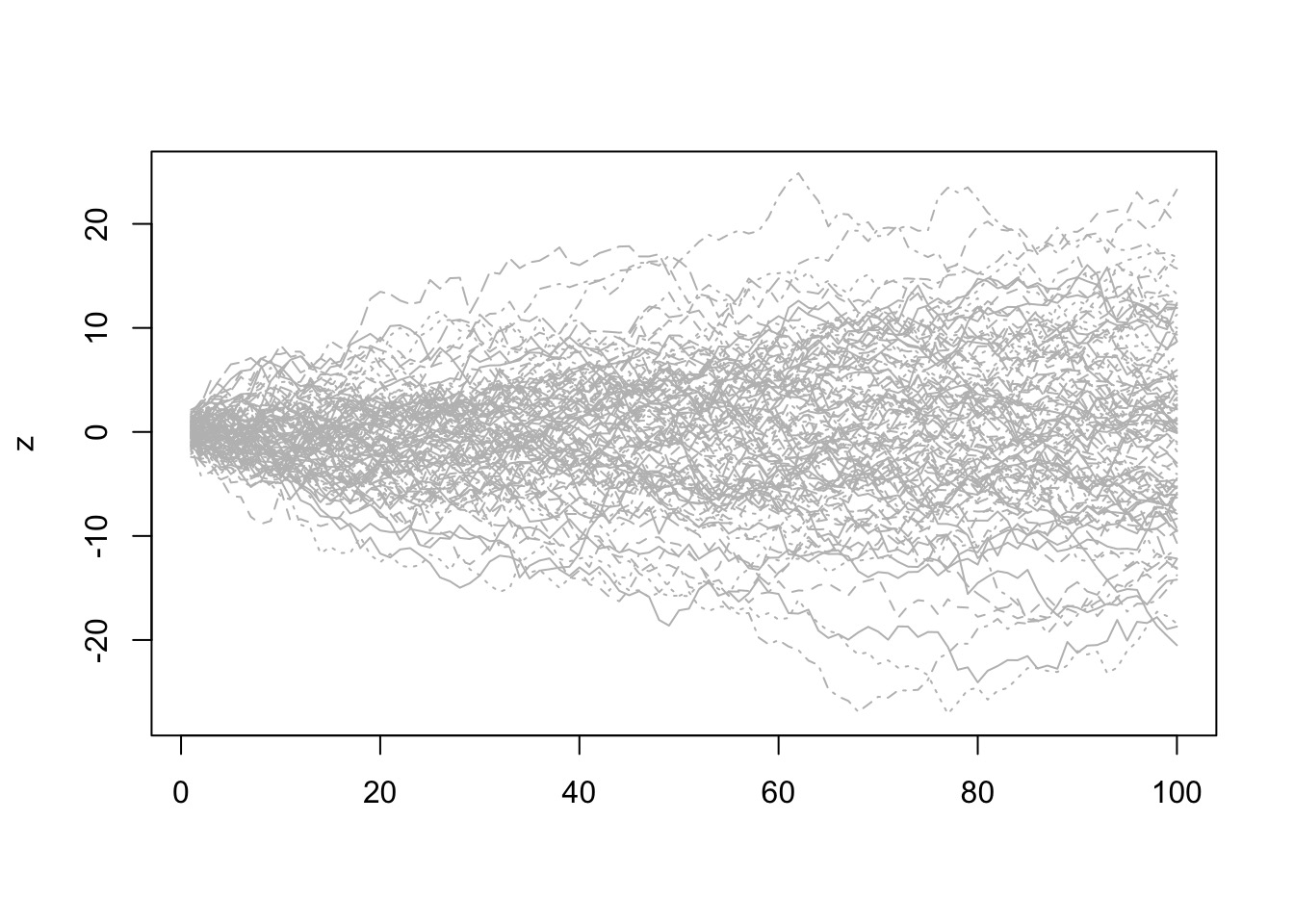
A Normal Error Distribution and Many Small Errors
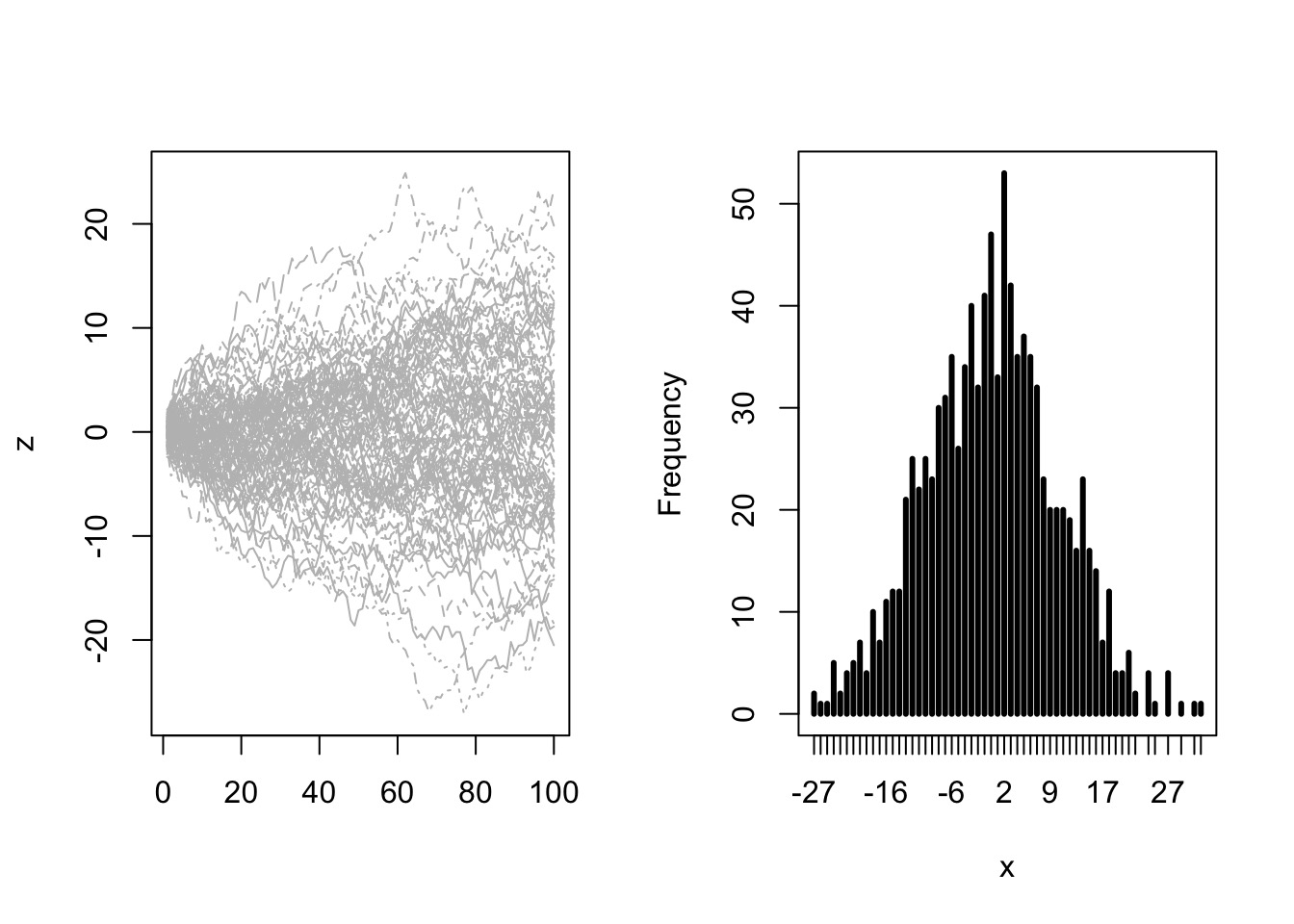
Or in Continuous Space
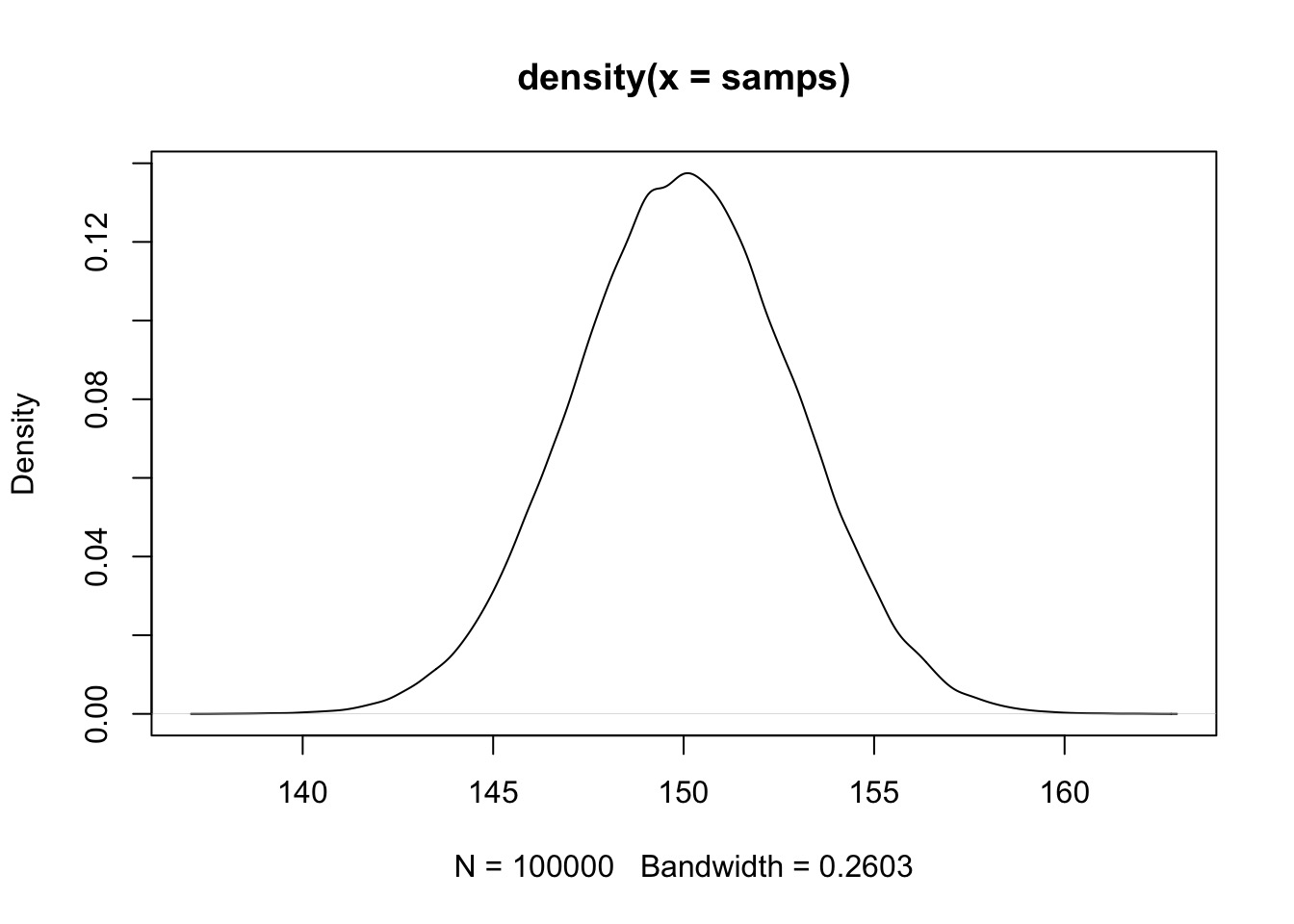
Gaussian Behavior from Many Distributions
Code
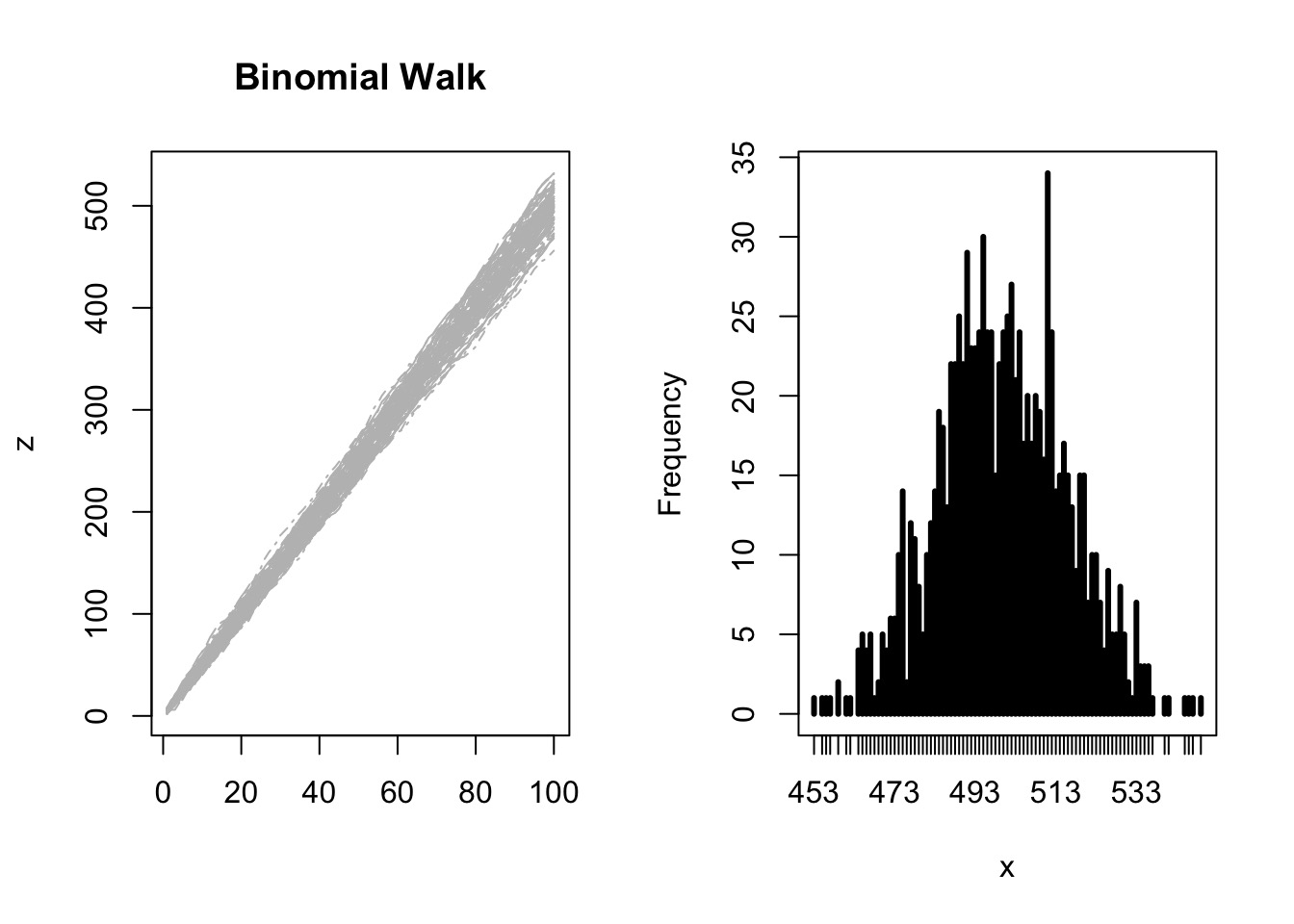
Try it: the Central Limit Theorem
But, Is the World an Accumulation of Small Errors?
Linear Regression: A Simple Statistical Golem
- Describes association between predictor and response
- Response is additive combination of predictor(s)
- Constant variance
Why should we be wary of linear regression?

- Approximate
- Not mechanistic
- Often deployed without thought
- But, often very accurate
So, how do we build models?
Identify response
Determine likelihood (distribution of error of response)
Write equation(s) describing generation of predicted values
Assign priors to parameters
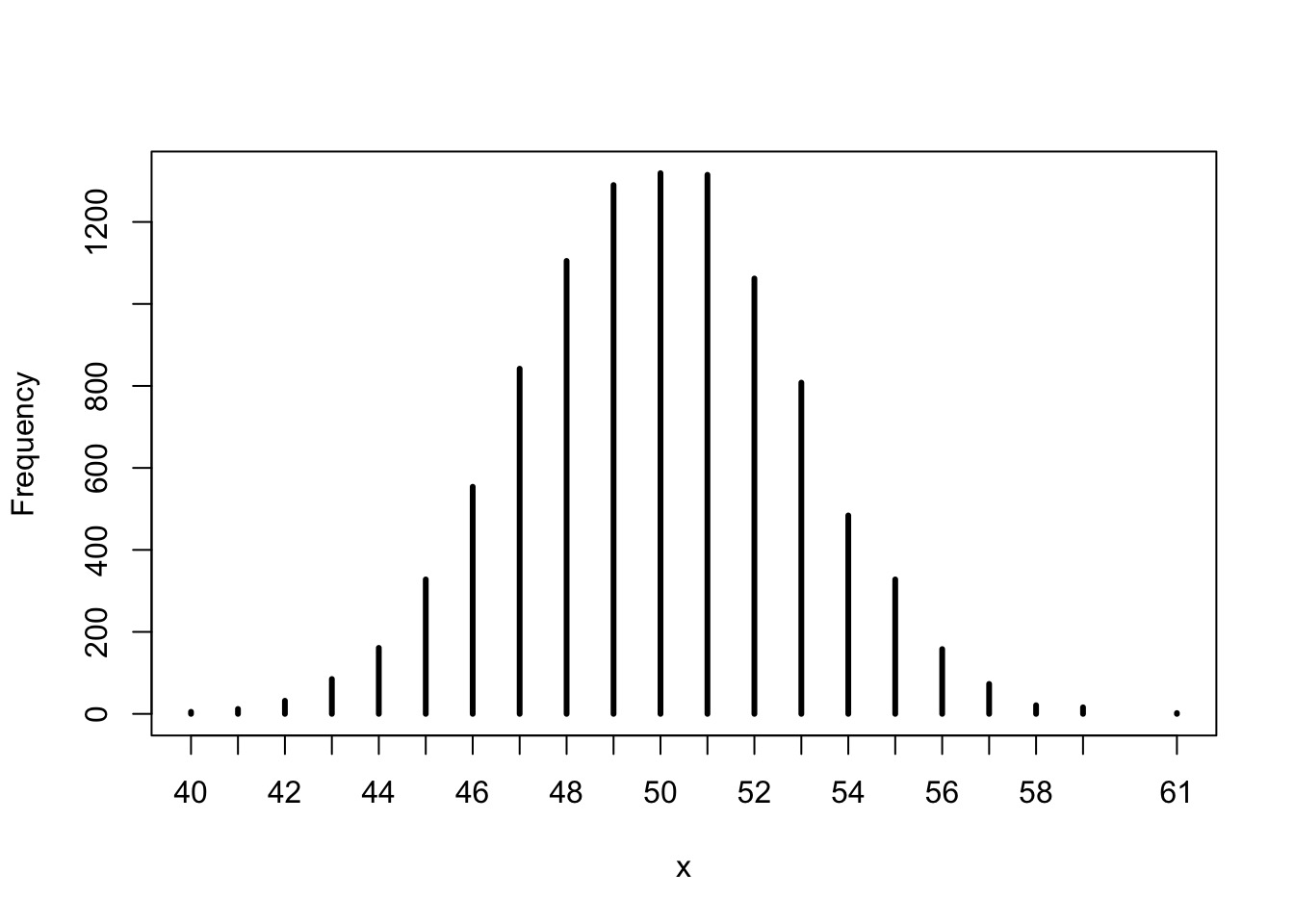
Our Previous Model
Likelihood:
\(w \sim Binomial(6, size=9, prob = prob)\)
Prior:
\(prob \sim Uniform(0,1)\)
A Normal/Gaussian Model
Likelihood:
\(y_i \sim Normal( \mu, \sigma)\)
Prior:
\(\mu \sim Normal(0,1000)\)
\(\sigma \sim U(0,50)\)
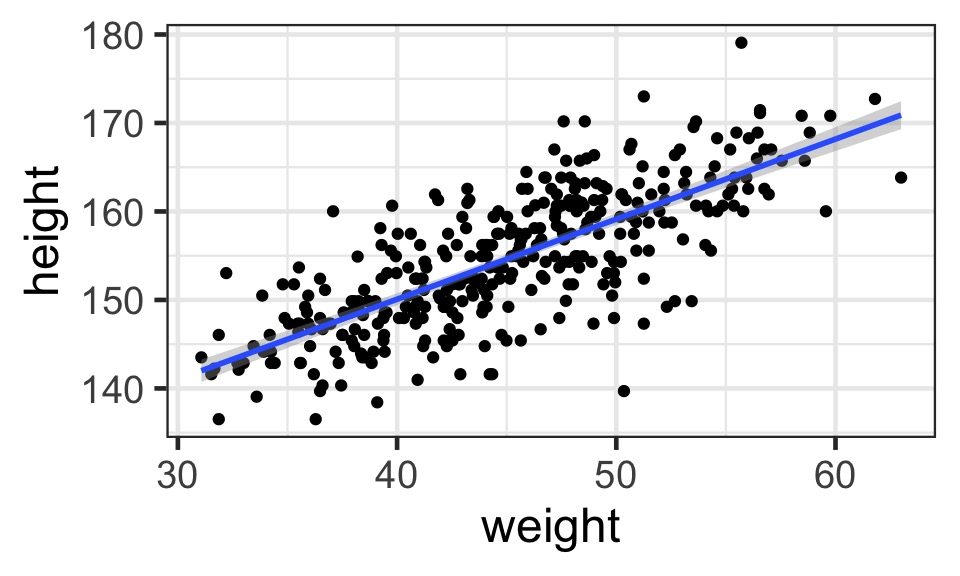
A Model of a Mean Height from the !Kung San

What does the data look like?
Our Model of Height
Likelihood:
\(h_i \sim Normal( \mu, \sigma)\)
Prior:
\(\mu \sim Normal(187, 20)\) My Height
\(\sigma \sim U(0,50)\) Wide range of possibilities
Priors
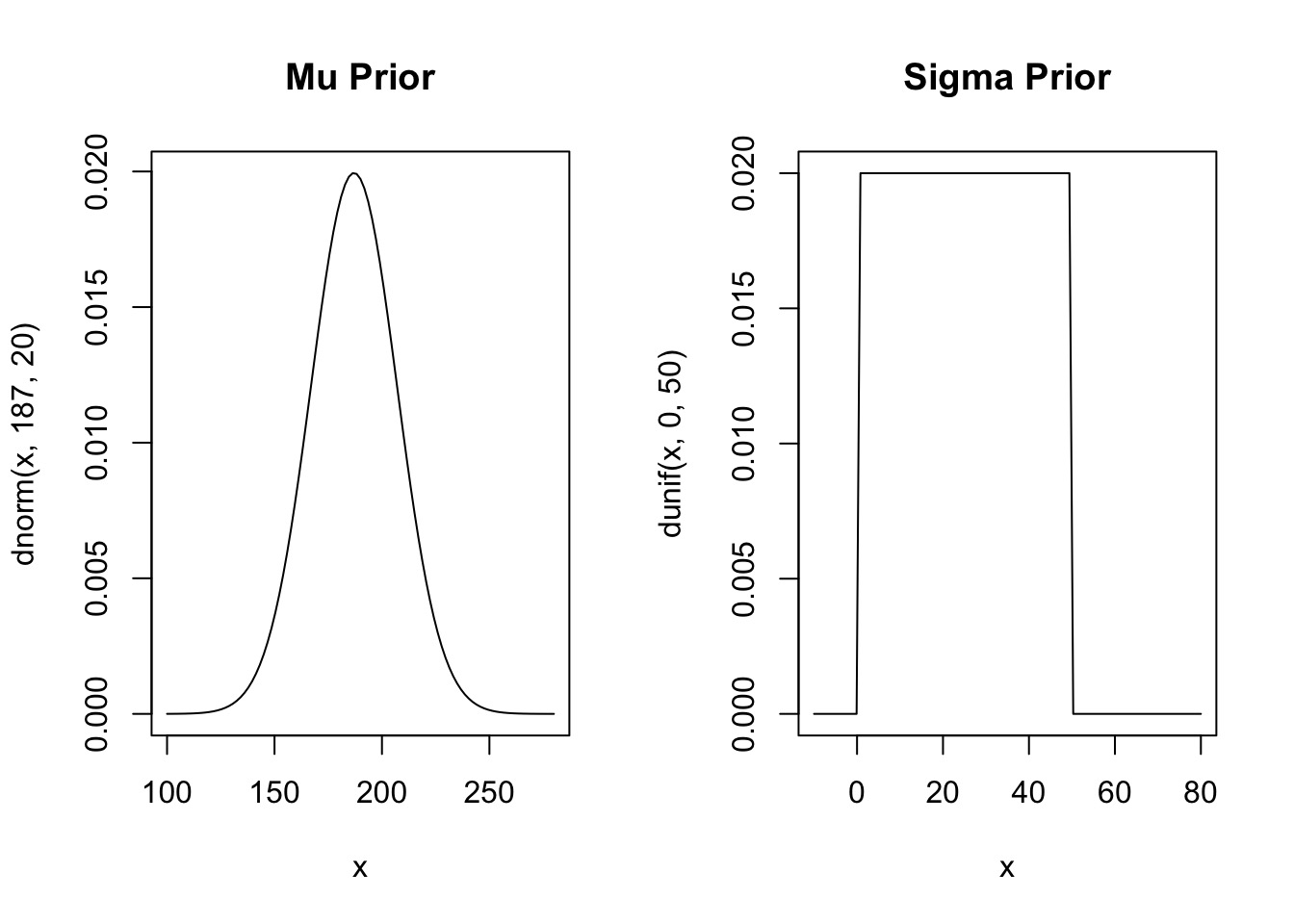
Prior Predictive Simulation
Are your priors any good?
Simulate from them to generate fake data
Does simulated data look realistic?
Does simulated data at least fall in the range of your data?
Example:
Prior:
\(\mu \sim \mathcal{N}(187, 20)\) My Height
\(\sigma \sim \mathcal{U}(0,50)\) Wide range of possibilities
Reasonable? Giants and Negative People?
Prior:
\(\mu \sim \mathcal{N}(187, 20)\) My Height
\(\sigma \sim \mathcal{U}(0,50)\) Wide range of possibilities
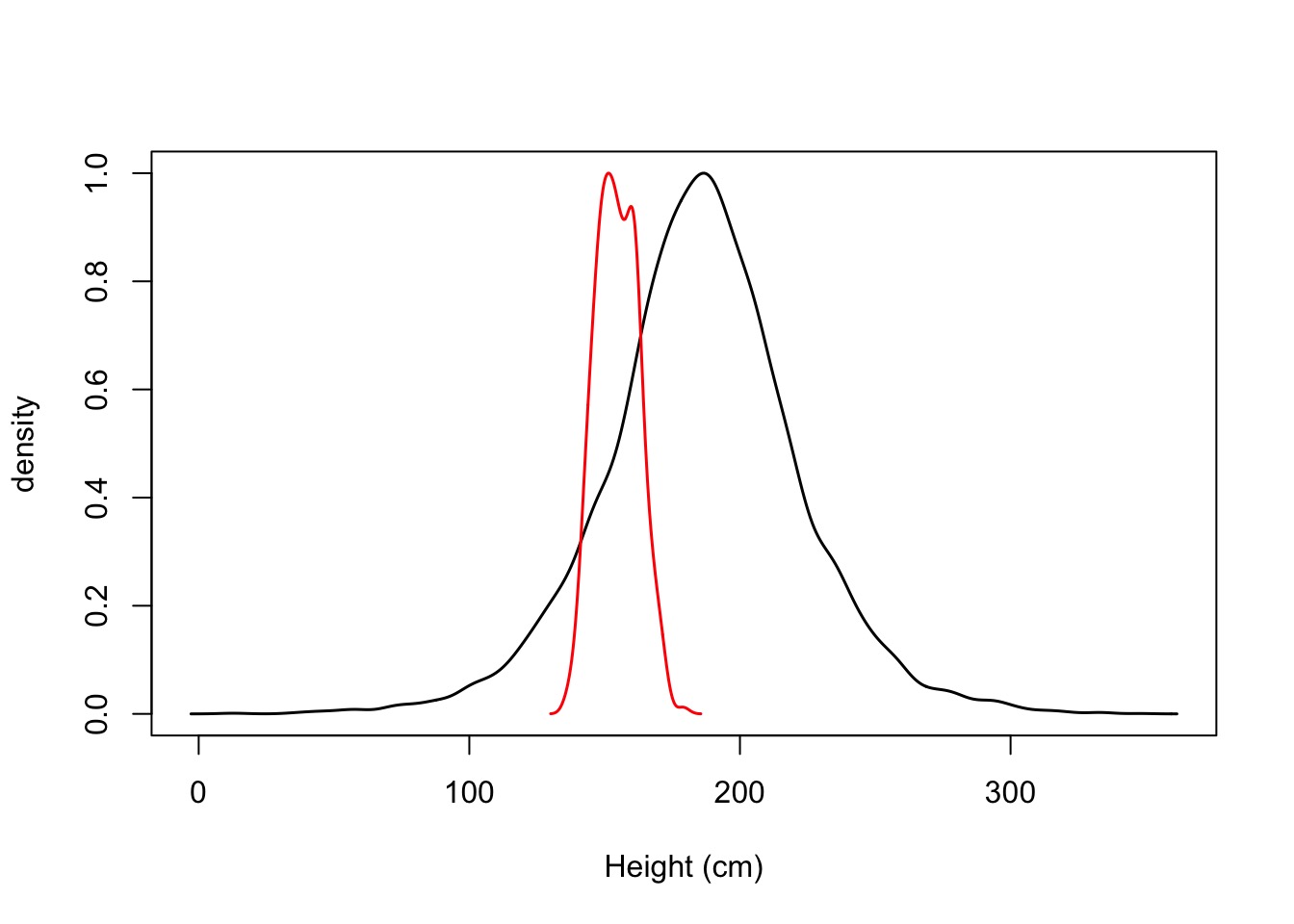
OK, I’m Tall, And Not Even In the Data, So….
Prior:
\(\mu \sim \mathcal{N}(150, 20)\) Something More Reasonable
\(\sigma \sim \mathcal{U}(0,50)\) Wide range of possibilities
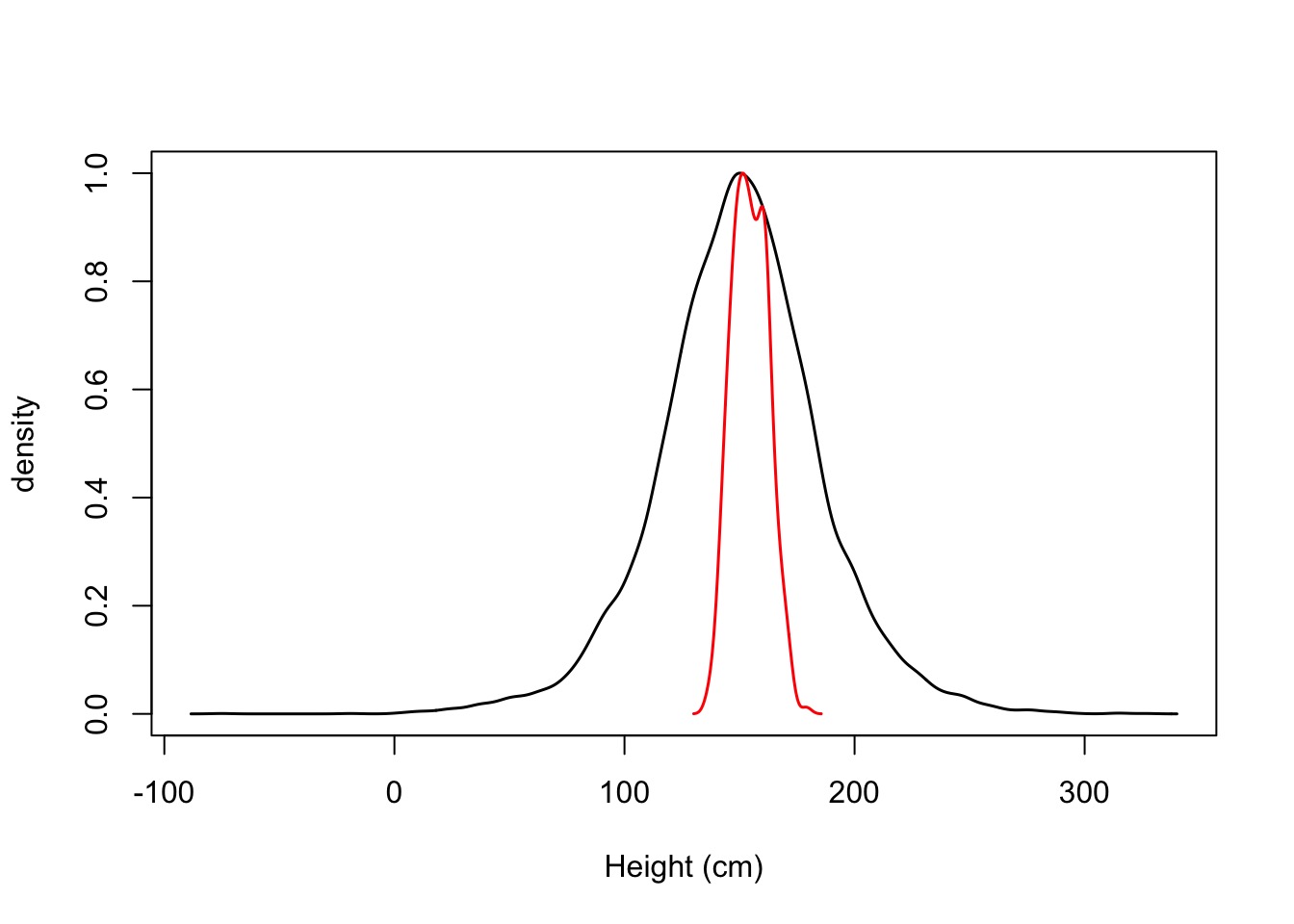
Grid Sampling
# Make the grid
grid <- tidyr::crossing(mu = seq(140, 160, length.out=200),
sigma = seq(4, 9, length.out=200)) |>
#Calculate the log-likelihoods for each row
group_by(1:n()) %>%
mutate(log_lik = sum(dnorm(Howell1_Adult$height, mu, sigma, log=TRUE))) %>%
ungroup() %>%
# Use these and our posteriors to get the numerator
# of Bayes theorem
mutate(numerator = log_lik +
dnorm(mu, 150, 20, log=TRUE) +
dunif(sigma, 0,50, log=TRUE)) |>
#Now calculate the posterior (approximate)
mutate(posterior = exp(numerator - max(numerator)))Posterior
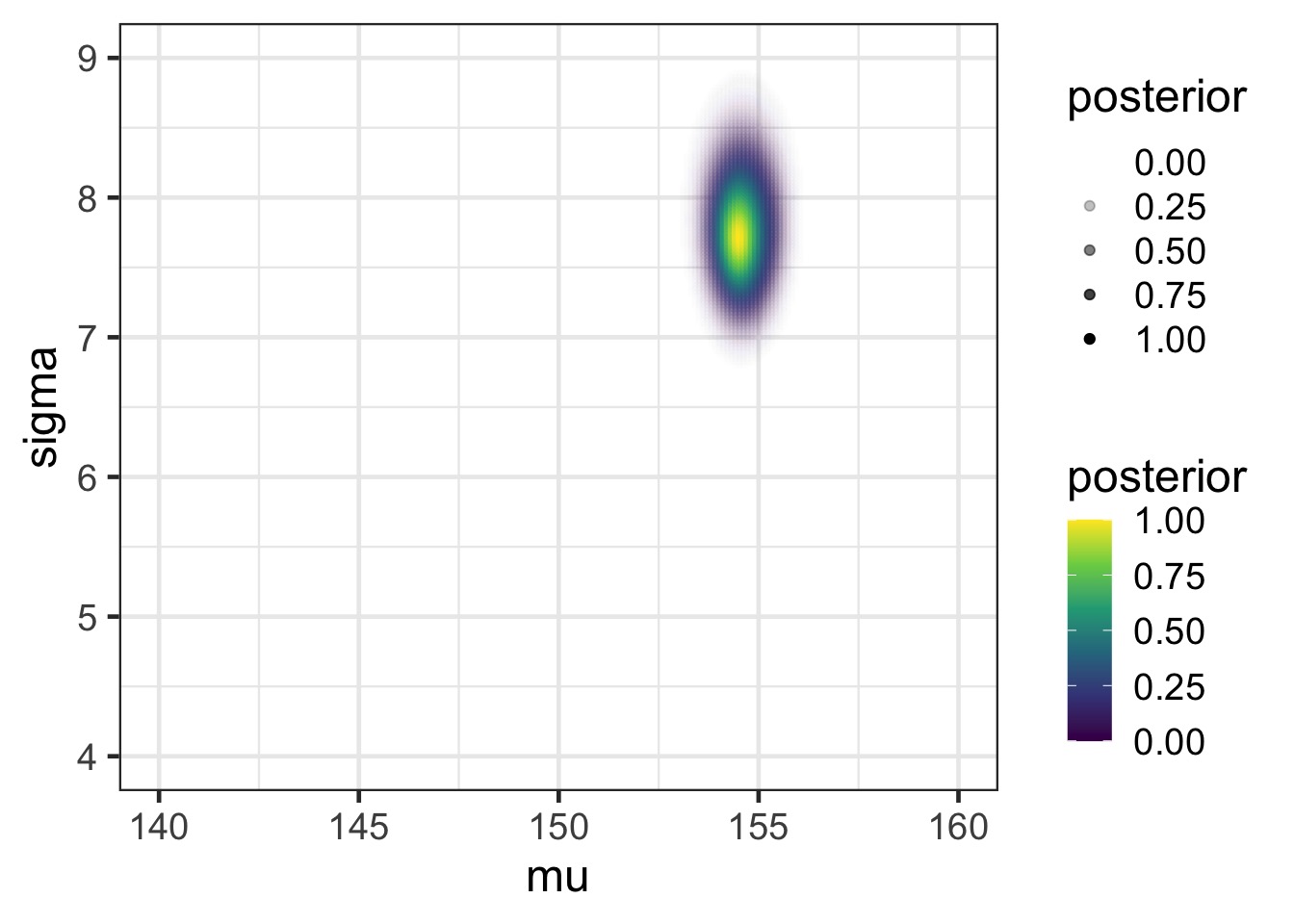
Posterior from a Sample of the Grid

Or, let’s Reconceptualize With a Model
Likelihood:
\(h_i \sim \mathcal{N}( \mu, \sigma)\) height ~ dnorm(mu, sigma)
Prior:
\(\mu \sim \mathcal{N}(150, 20)\) mu ~ dnorm(150, 200)
\(\sigma \sim U(0,50)\) sigma ~ dunif(0,50)
Building Models using rethinking: The alist Object
Building Models using rethinking: The alist Object
Feed the Model to Maximum A Posterior Approximation
- Uses optimization algorithms
- Same algorithms as likelihood
Compare map to grid
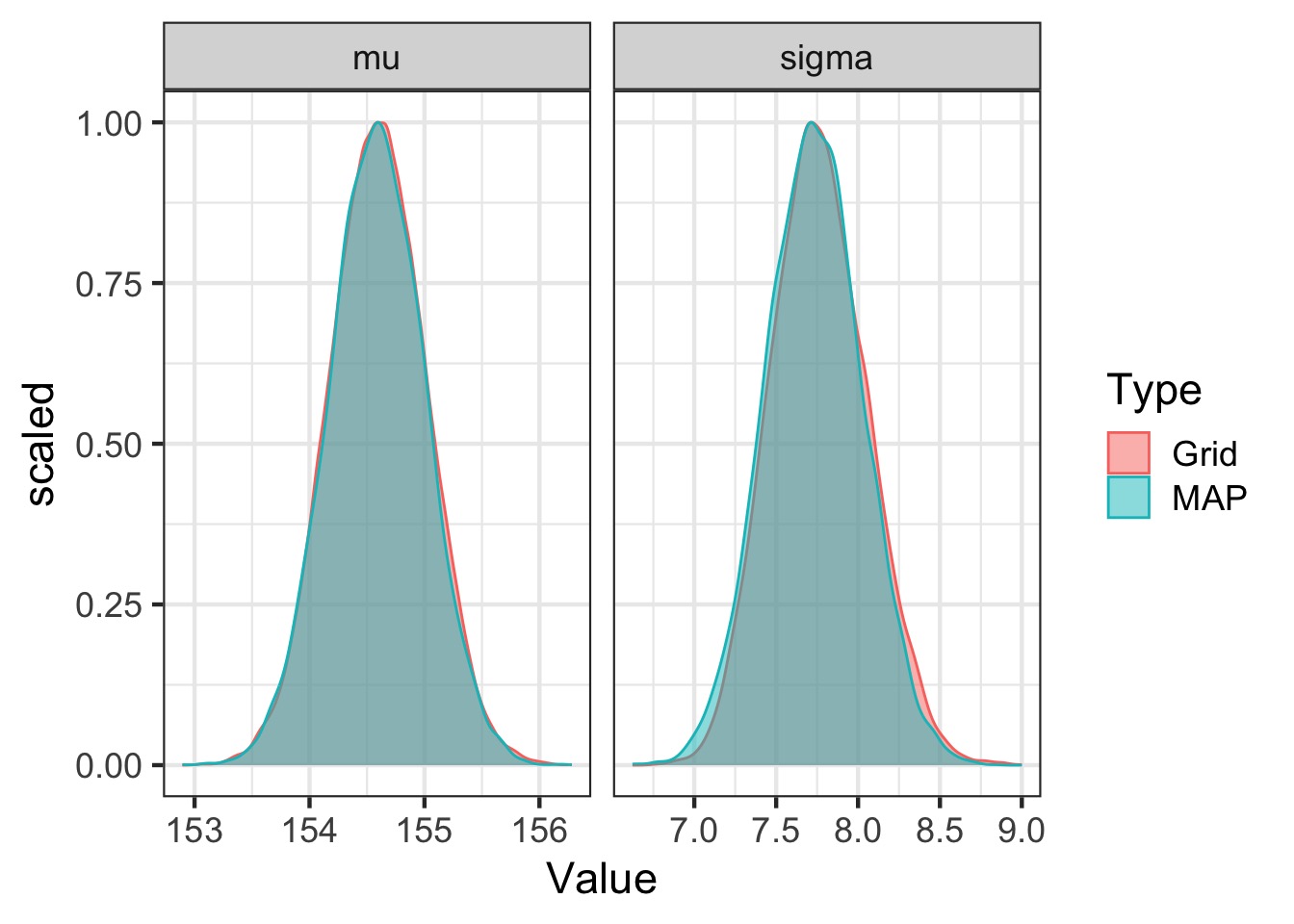
Adding a Predictor
Identify response (height)
Determine likelihood (distribution of error of response)
Write equation(s) describing generation of predicted values
- Weight predicts height
Assign priors to parameters
- Check priors with simulation
The Mean Changes with predictor: A Linear Model!
Likelihood:
\(h_i \sim \mathcal{N}(\mu_i, \sigma)\)
Data Generating Process
\(\mu_i = \alpha + \beta (x_i - \bar{x_i})\)
Prior:
\(\alpha \sim \mathcal{N}(150, 20)\) Previous Mean
\(\beta \sim \mathcal{N}(0, 10)\) Weakly Informative
\(\sigma \sim \mathcal{U}(0,50)\) Wide range of possibilities
Let’s have the Centering Talk. Why?
Let’s Check our Priors! But Over what range?
So, ± 15
When in doubt, simulate it out!
set.seed(2019)
n <- 100
# a data frame of priors
sim_priors_df <- data.frame(a = rnorm(n, 150, 20),
b = rnorm(n, 0, 10))
# geom_abline to make lines
prior_plot <- ggplot(data = sim_priors_df) +
geom_abline(mapping = aes(slope = b, intercept = a)) +
xlim(c(-15,15)) + ylim(c(-100, 500)) +
xlab("Weight - Mean Weight") + ylab("Height (cm)") Giants and Negative Humans?
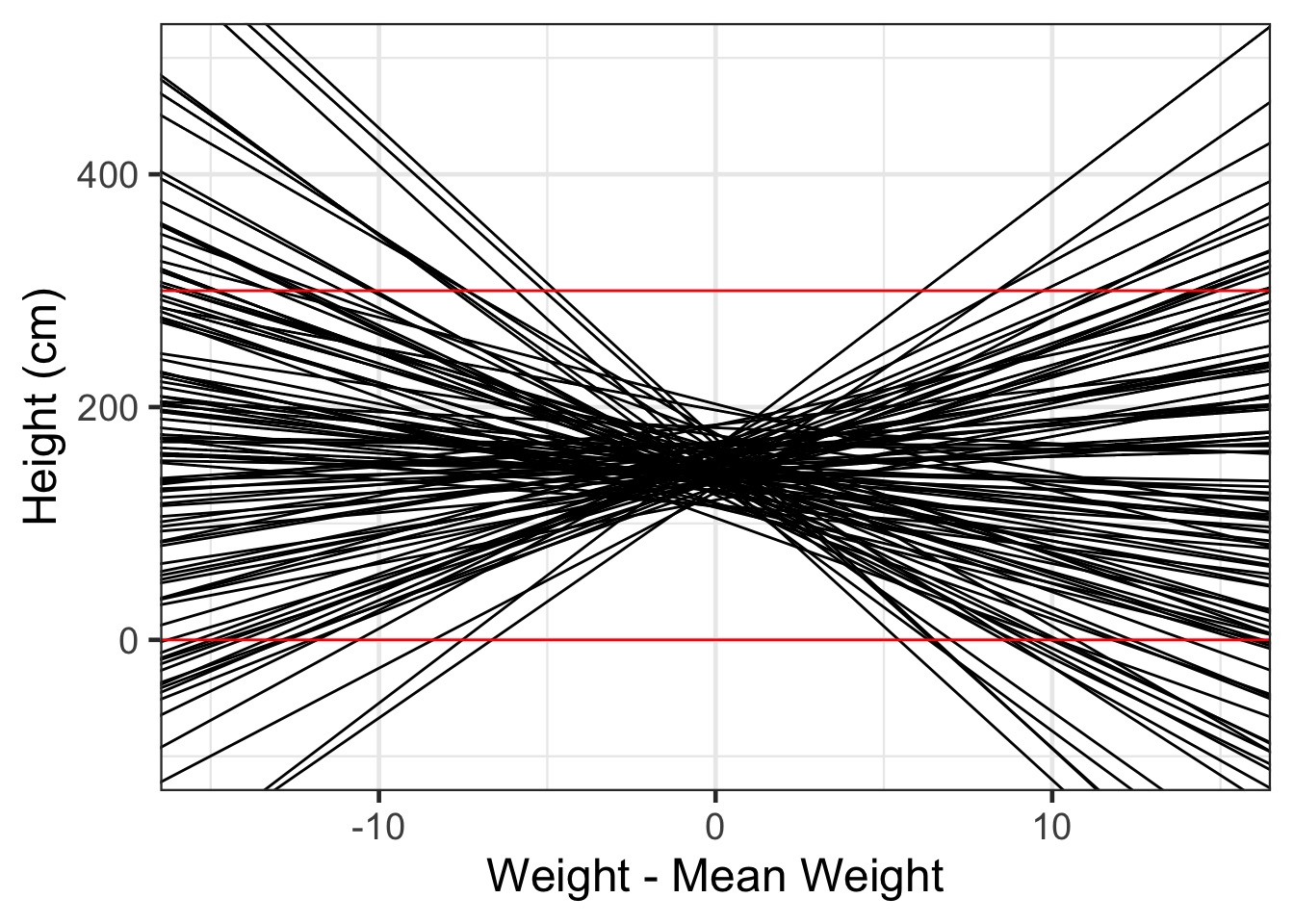
Rethinking our Priors!
Try a log-normal to guaruntee a positive value!
The Log Normal
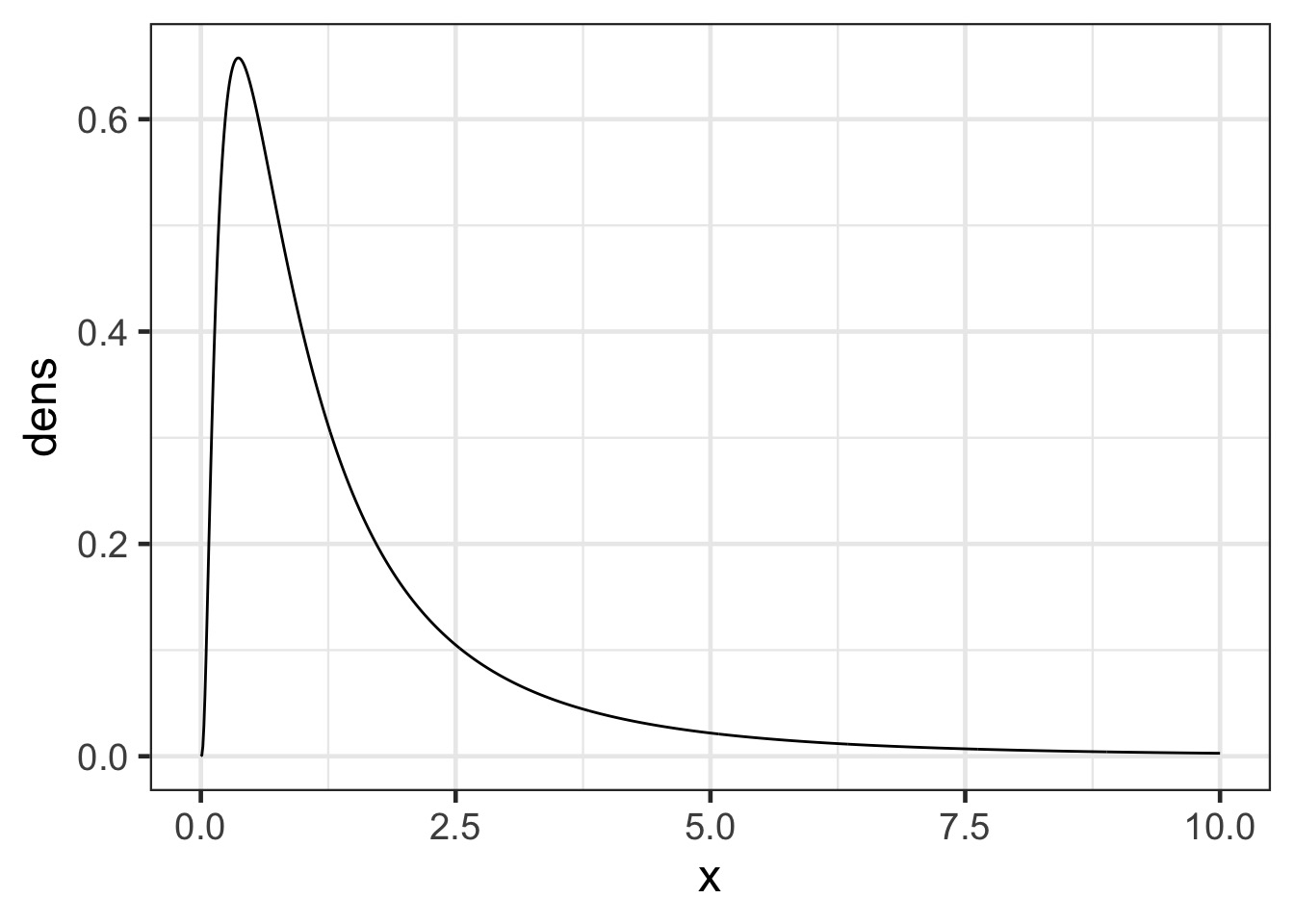
Simulate it out!
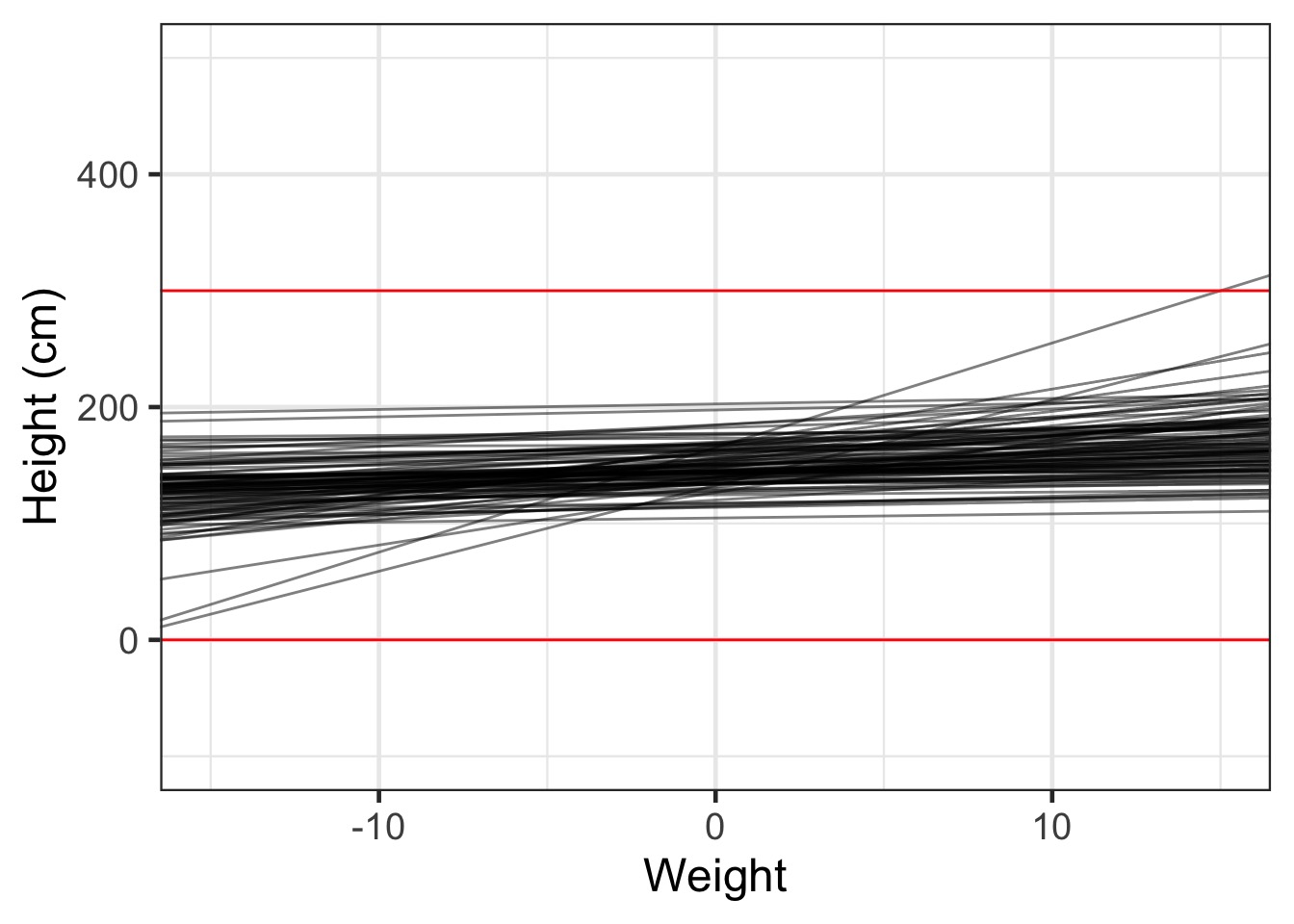
Our Linear Model
The Coefficients
See the fit
The fit
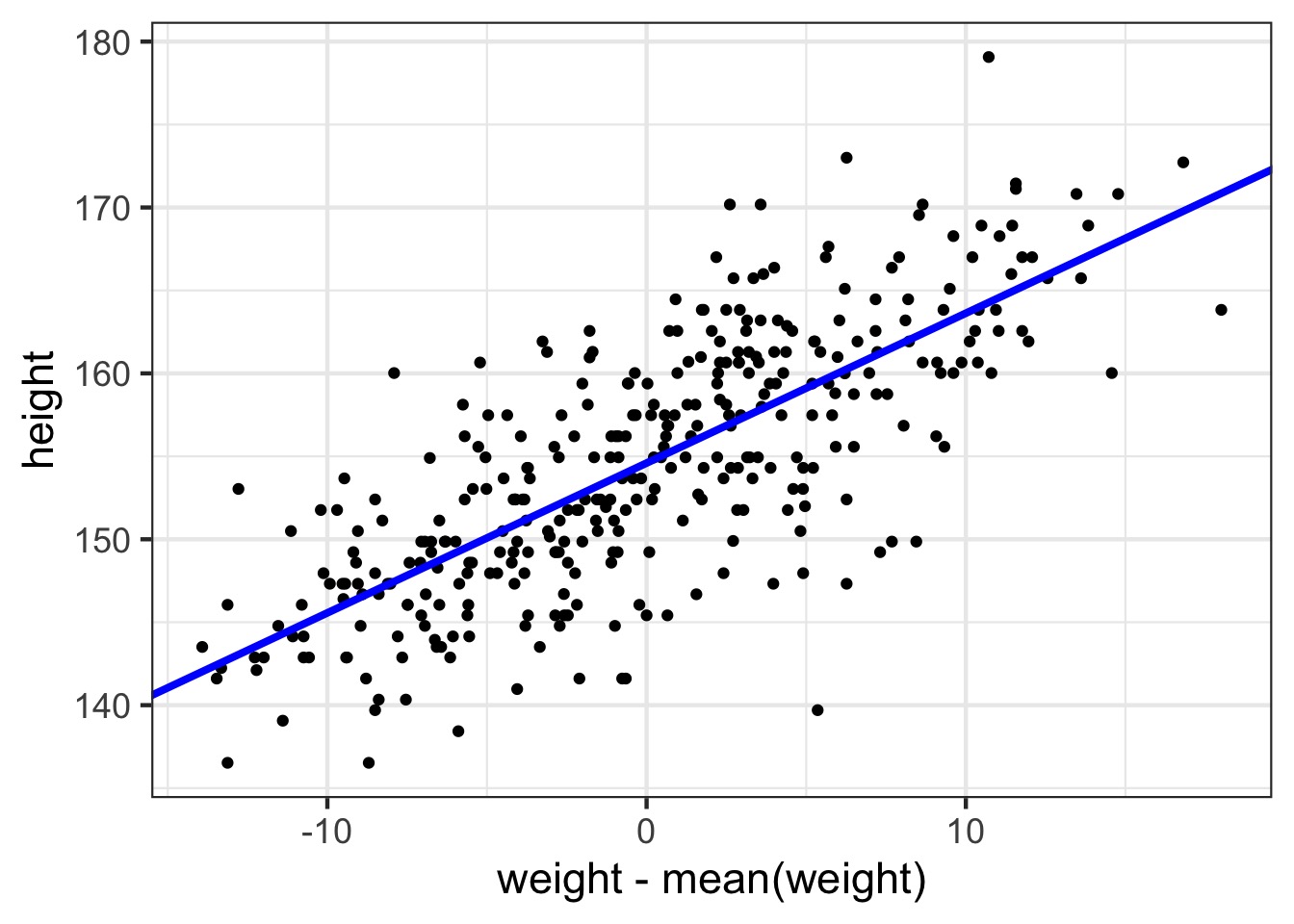
So, what do we do with a fit model?
Evaluate model assumptions
Evaluate model estimates and meaning
Assess uncertainty in fit
Assess uncertainty in prediction
Sampling From Models with tidybayes
We will use this as
tidybayesis great for many Bayesian Packages.Obeys a common tidy formatting
linpred_draws() for Link Predictions
weight_samps <-
linpred_draws(weight_fit,
newdata = Howell1_Adult,
ndraws = 1000)
head(weight_samps)# A tibble: 6 × 7
# Groups: height, weight, age, male, .row [1]
height weight age male .row .draw .value
<dbl> <dbl> <dbl> <int> <int> <int> <dbl>
1 152. 47.8 63 1 1 1 157.
2 152. 47.8 63 1 1 2 157.
3 152. 47.8 63 1 1 3 158.
4 152. 47.8 63 1 1 4 157.
5 152. 47.8 63 1 1 5 157.
6 152. 47.8 63 1 1 6 157.Get Residuals and a Summarized Frame
Check Posterior Against Data
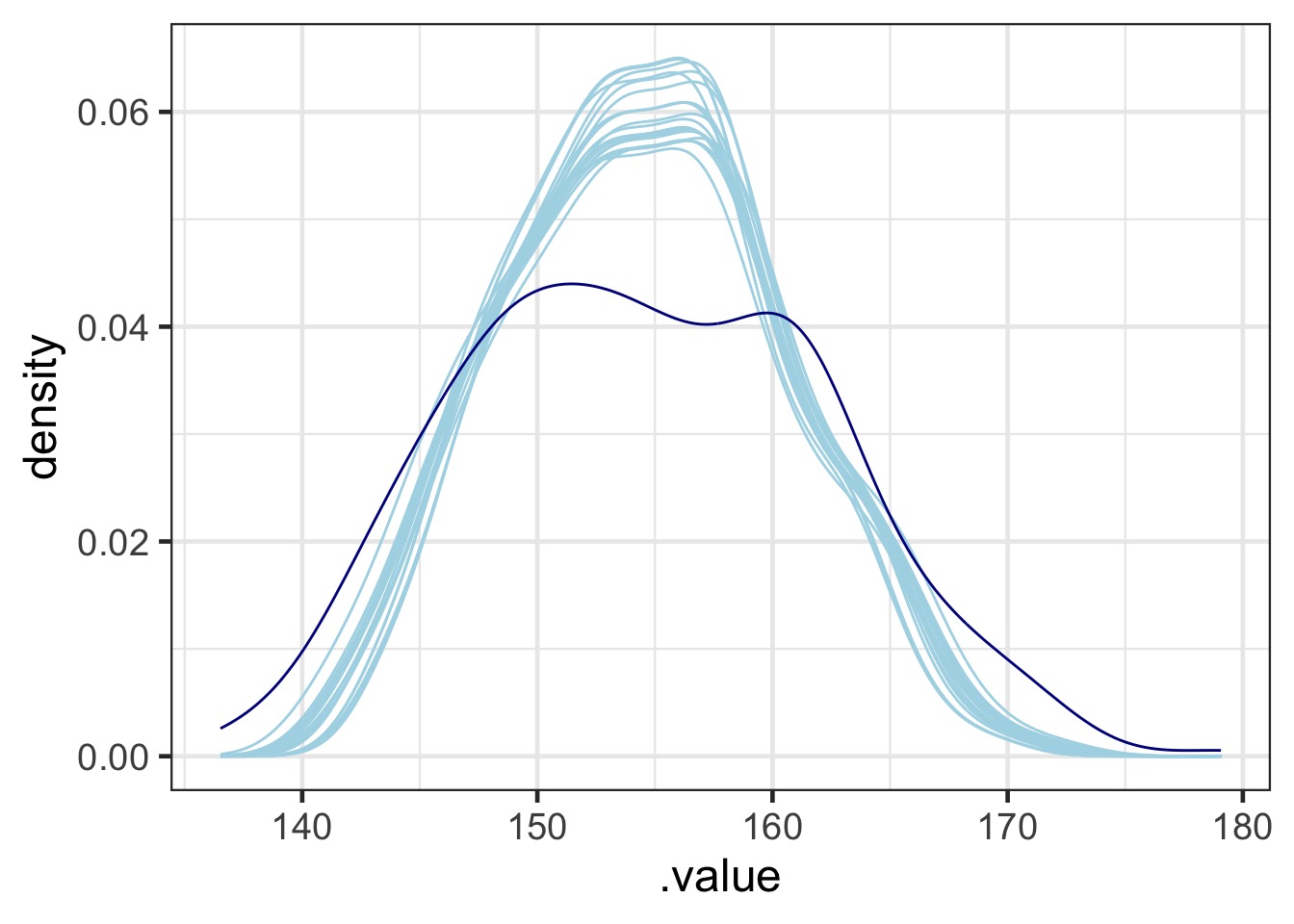
Code for PP Check
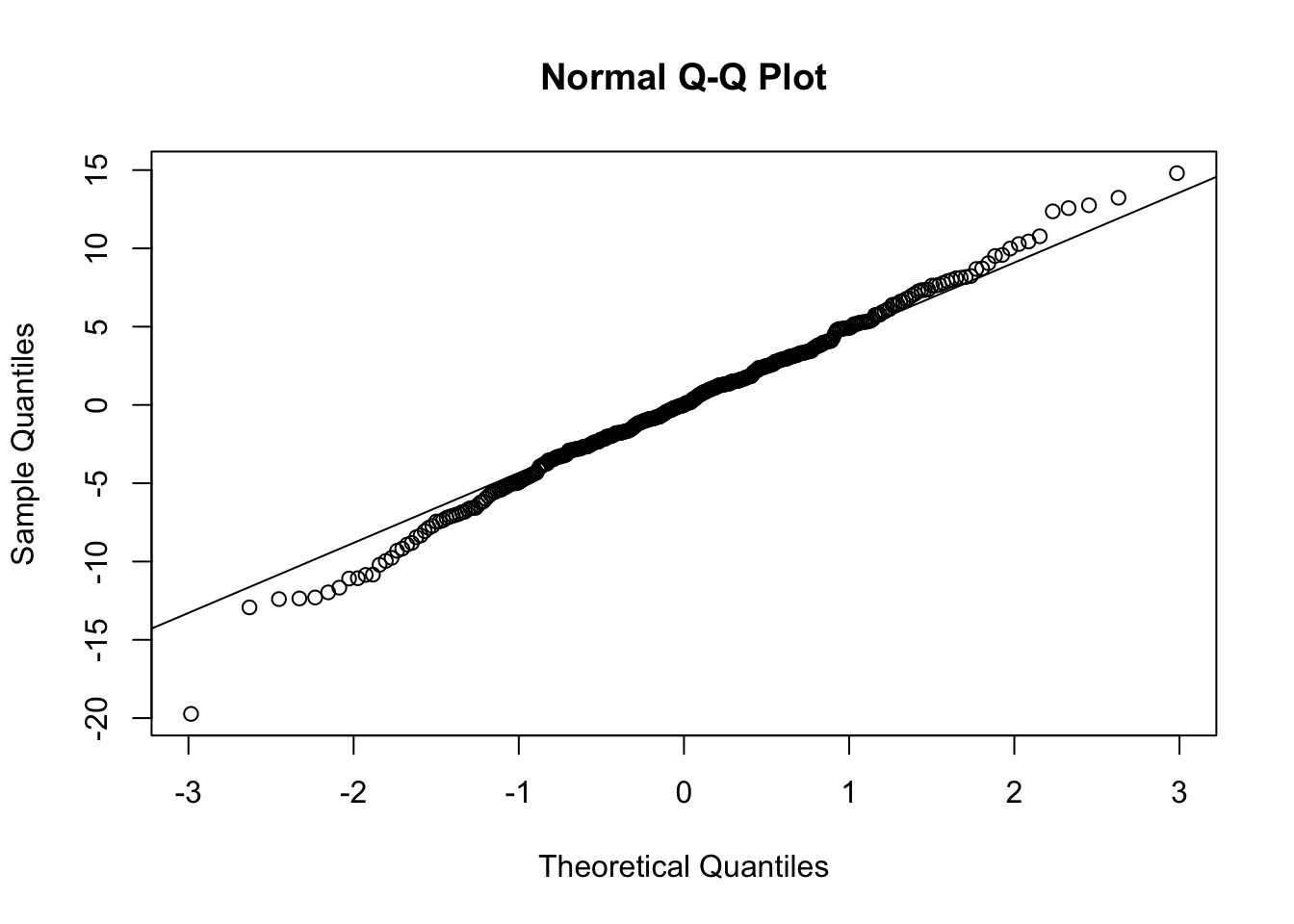
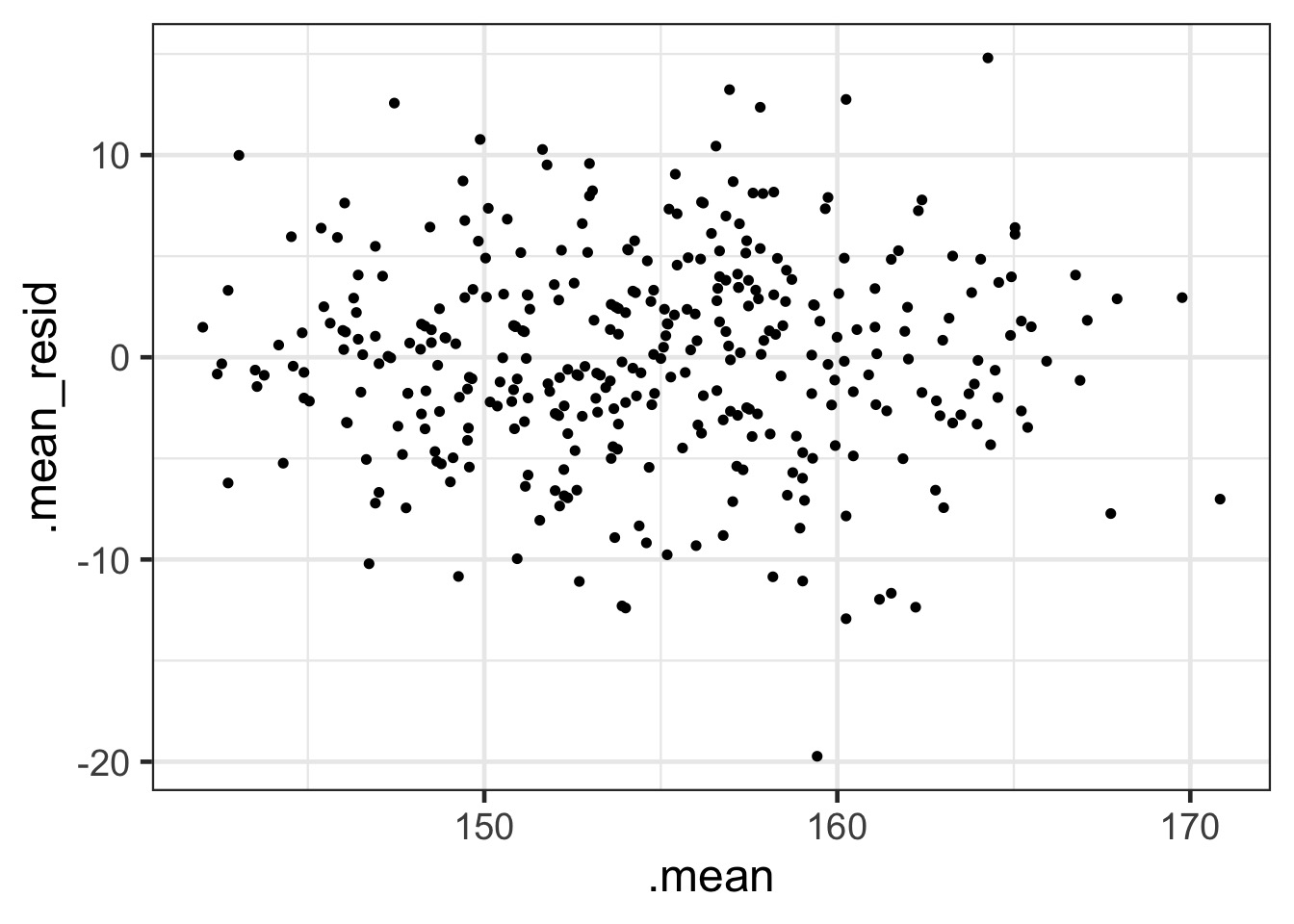
QQ, etc…
Fit-Residual
Code
Model Results
mean sd 5.5% 94.5%
alpha 154.5962531 0.27030757 154.16425 155.0282568
beta 0.9032821 0.04192365 0.83628 0.9702841
sigma 5.0718827 0.19115492 4.76638 5.3773852Are these meaningful?
Should you standardize predictors for a meaningful intercept?
Is this the right interval?
Model Results
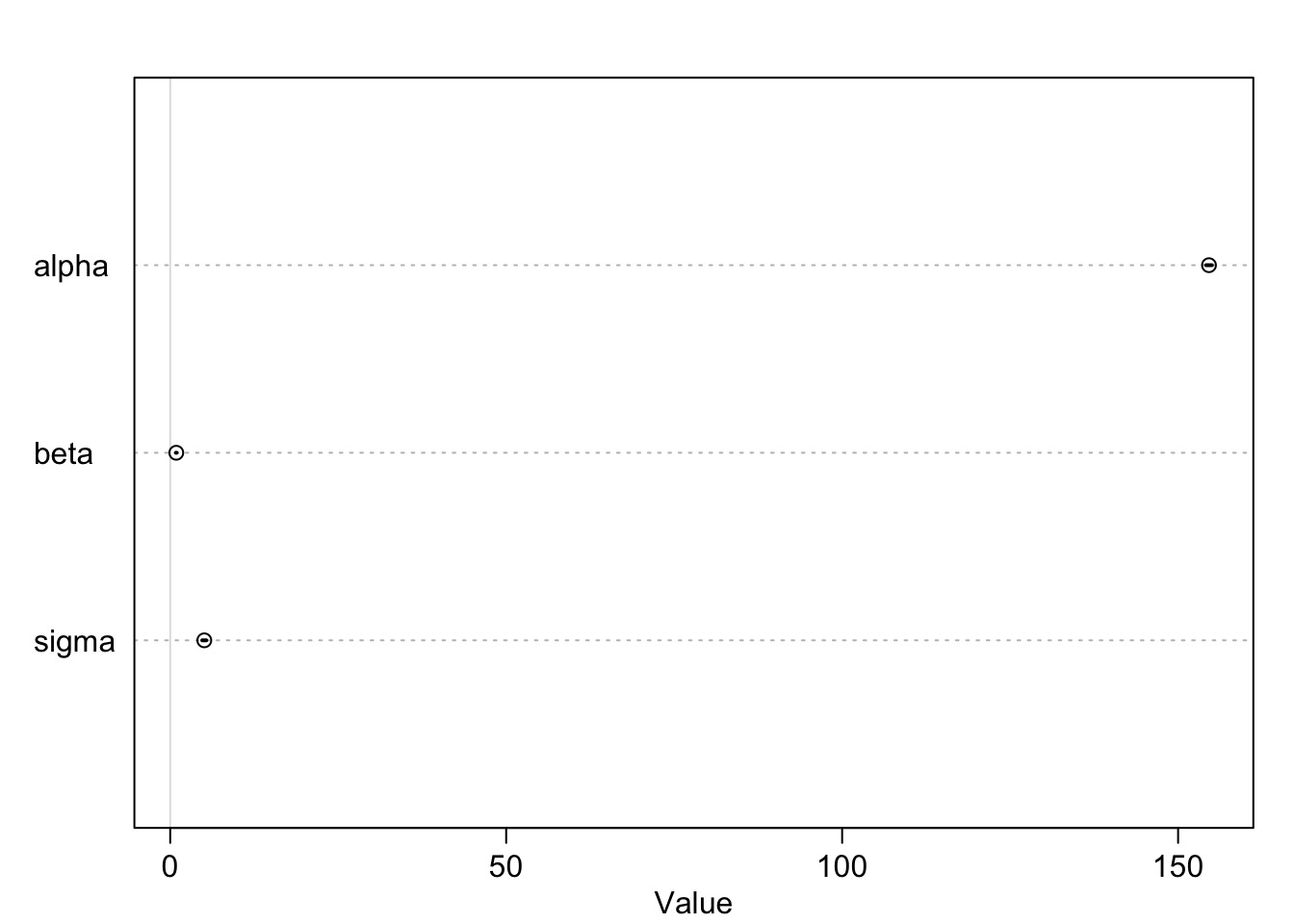
Sampling from the Posterior Distribution
Posteriors!
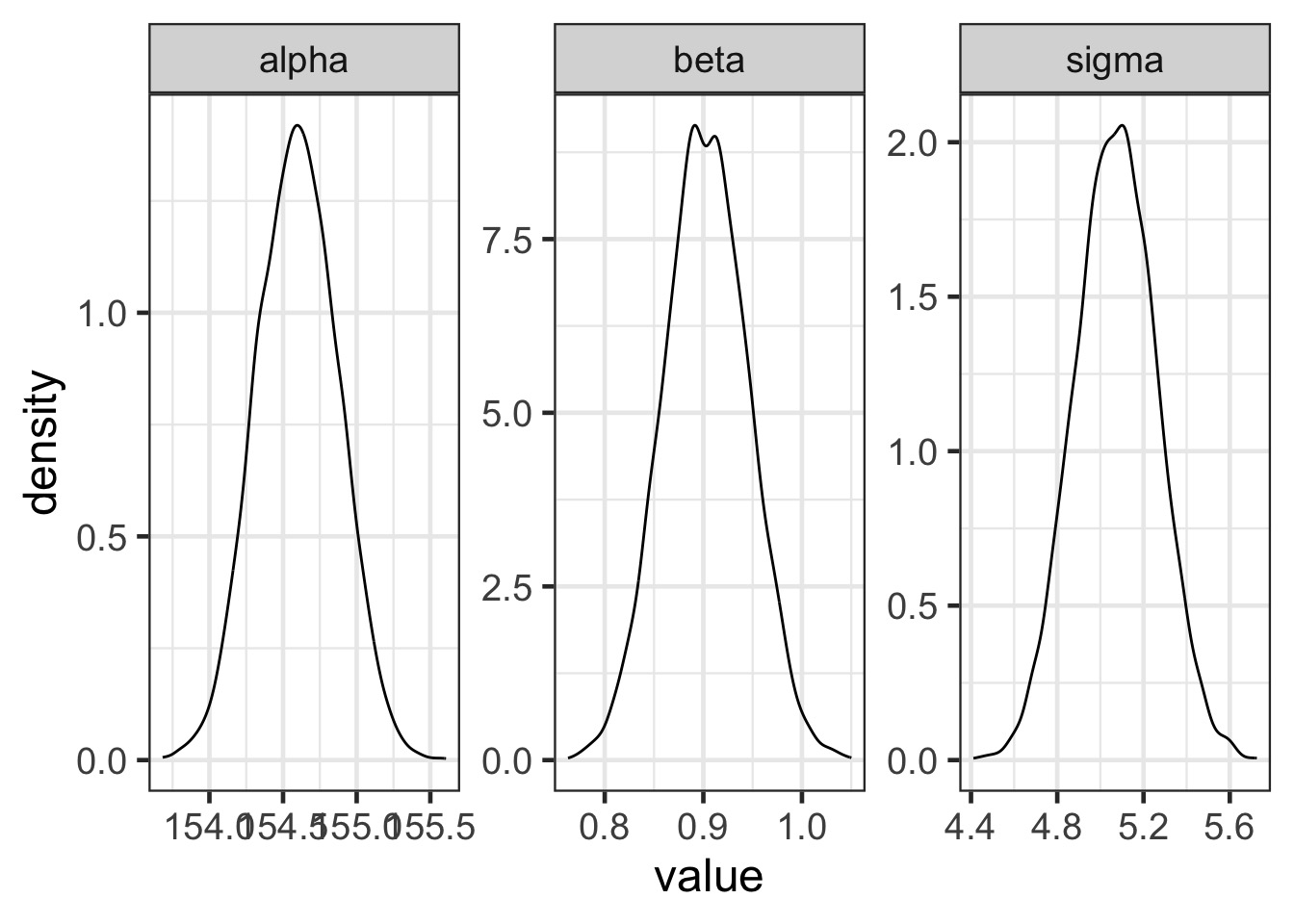
Code
Relationships?
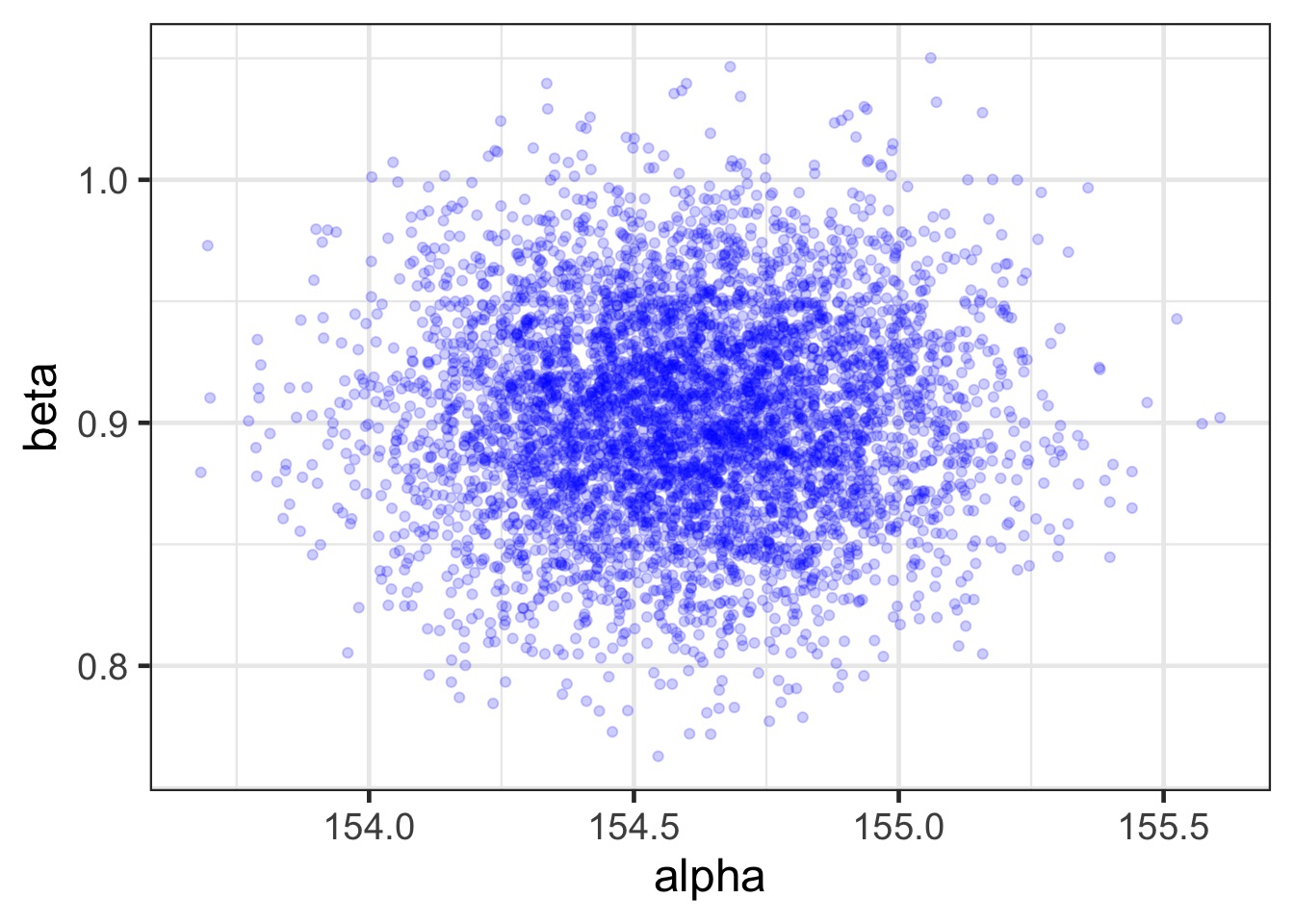
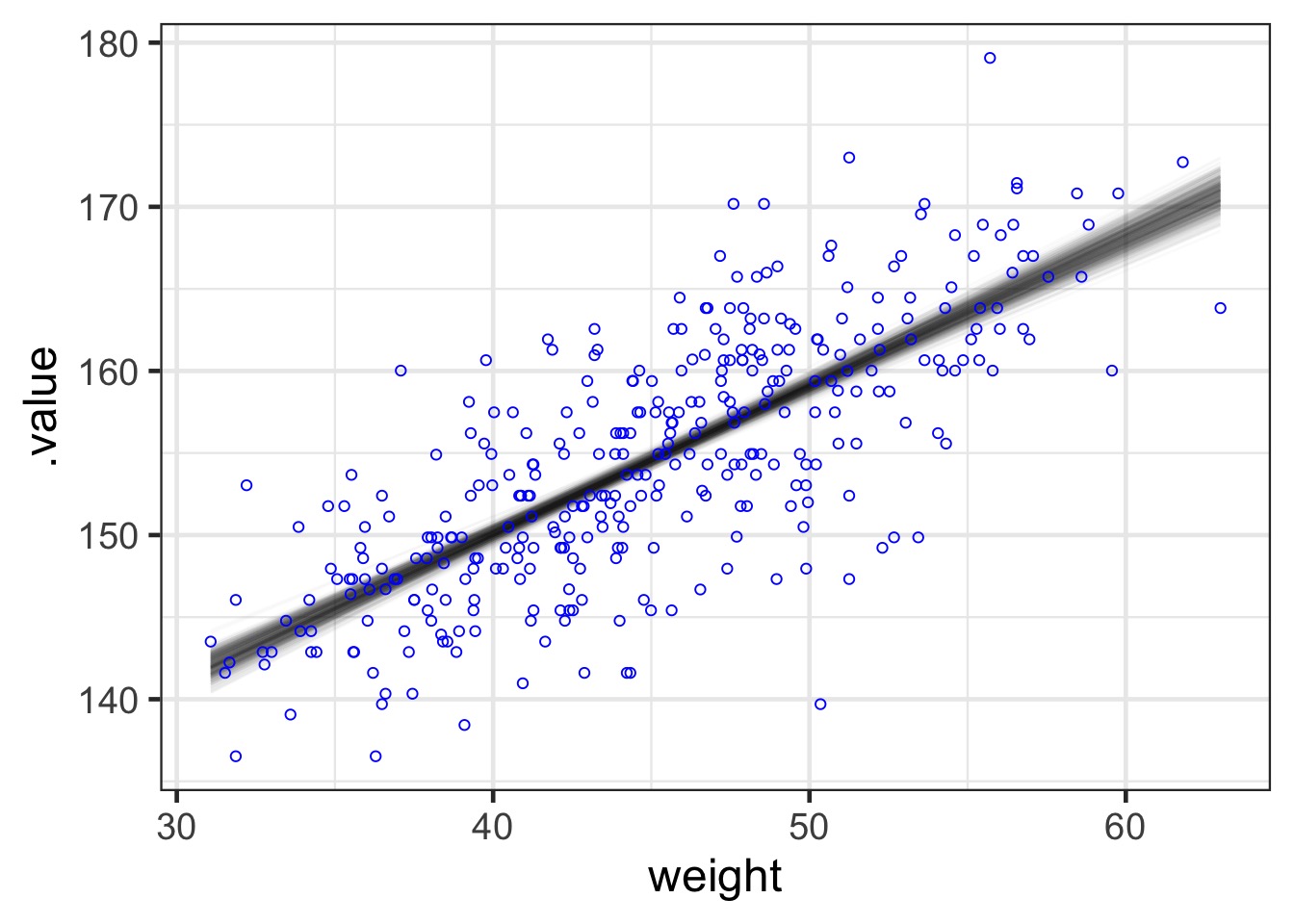
How Well Have we Fit the Data? Use Samples of Data
How Well Have we Fit the Data?
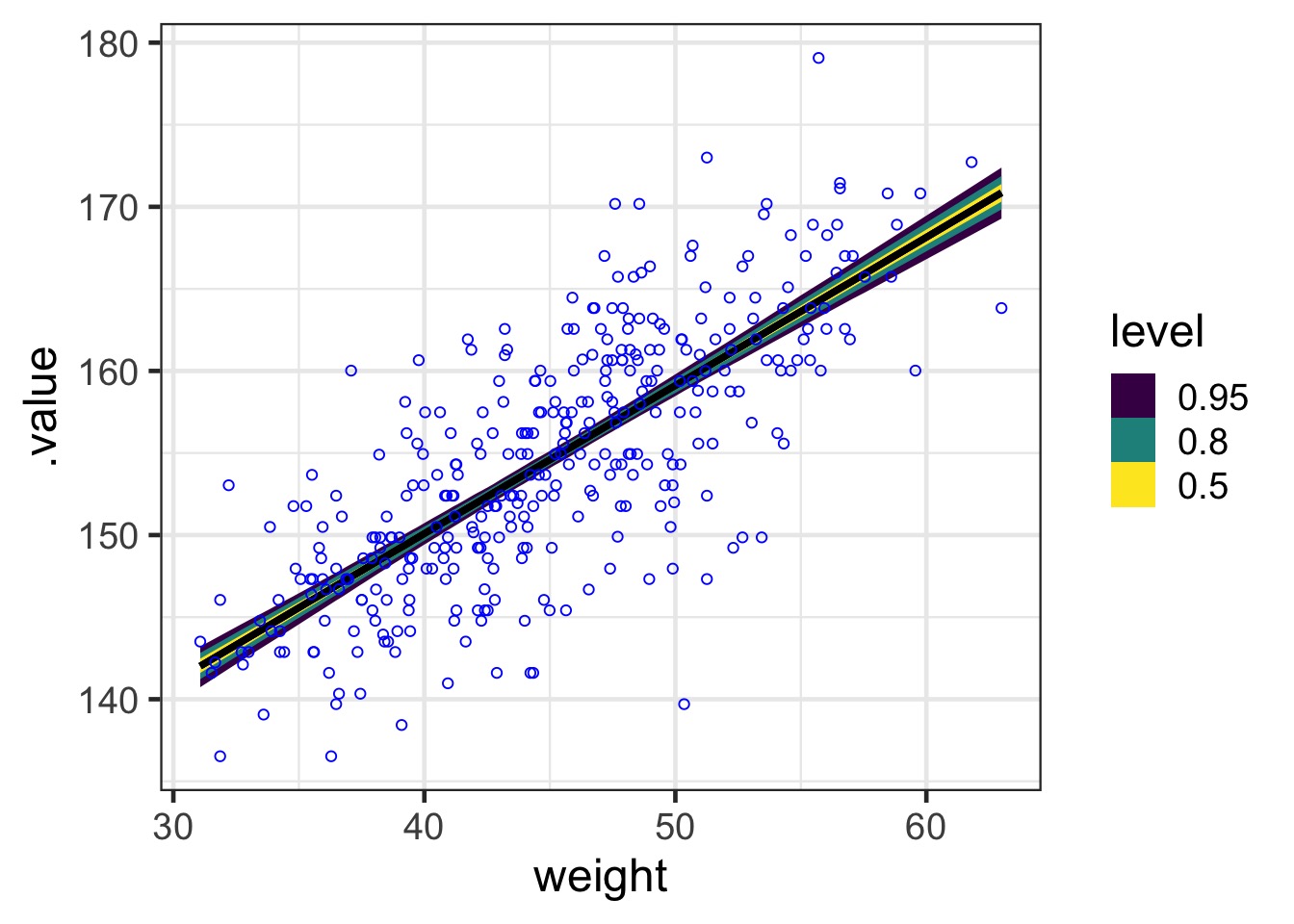
Or, Use ggdist
Or, Use ggdist
What about prediction intervals?
#1) Get the posterior prediction interval
pred_samps <- predicted_draws(weight_fit, newdata = Howell1_Adult)
#2) Put it all back together and plot
ggplot() +
#simulated fits from the posterior
stat_lineribbon(data=pred_samps ,
mapping=aes(x = weight,
y = .prediction)) +
#the data
geom_point(data=Howell1_Adult,
mapping=aes(x=weight, y=height), pch=1, color = "blue") Prediction Interval
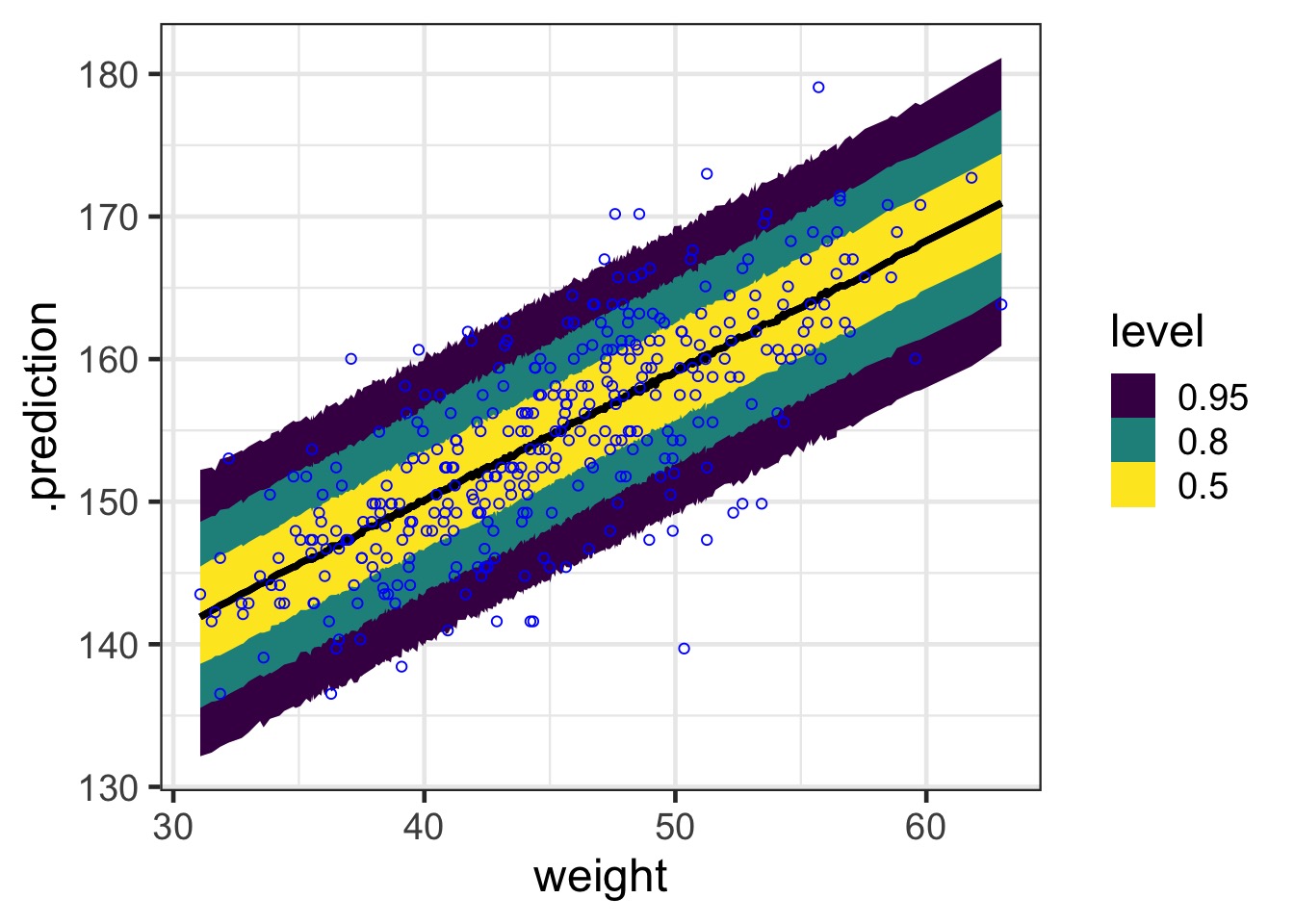
Prediction Interval Lines
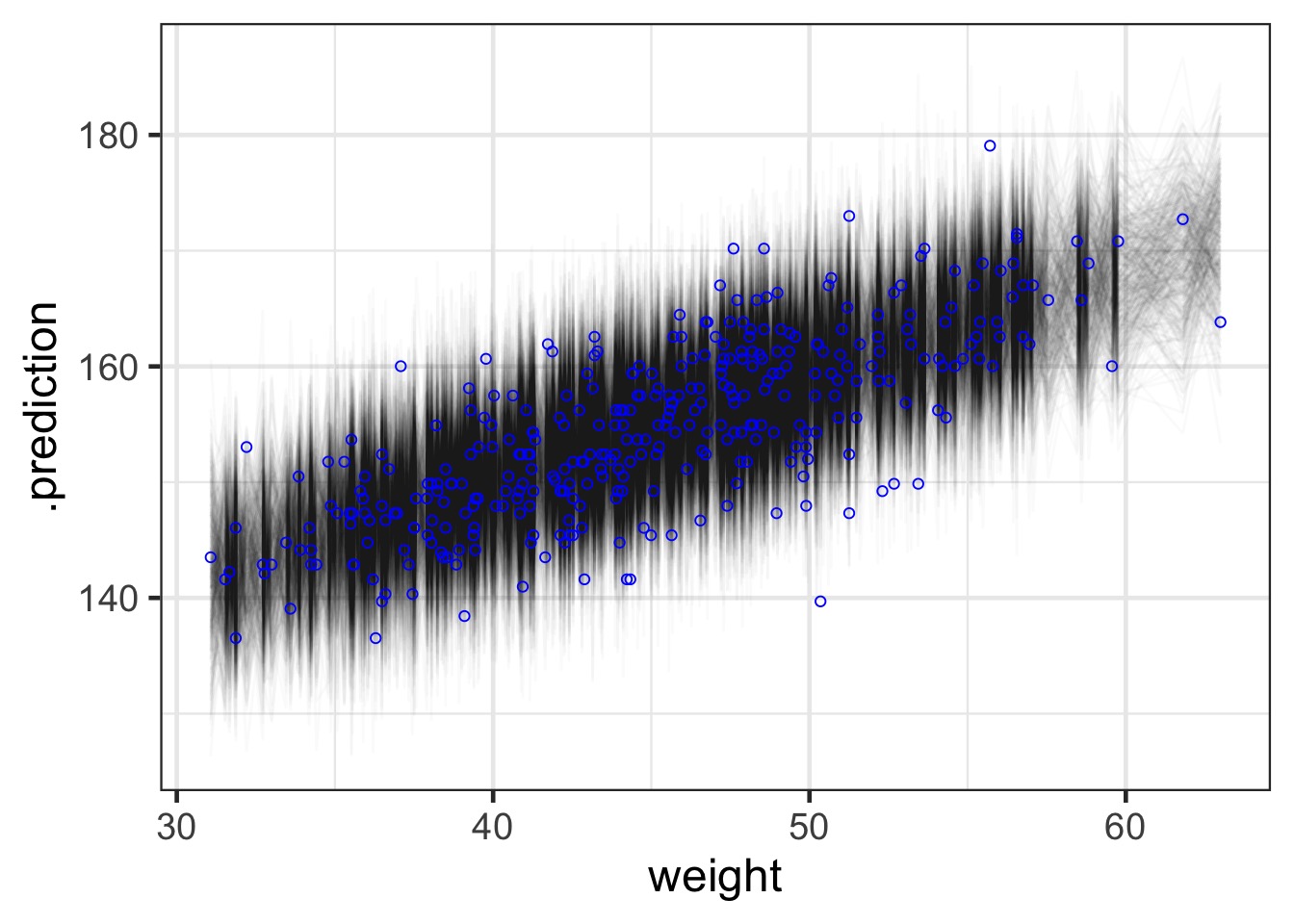
Prediction Interval Points
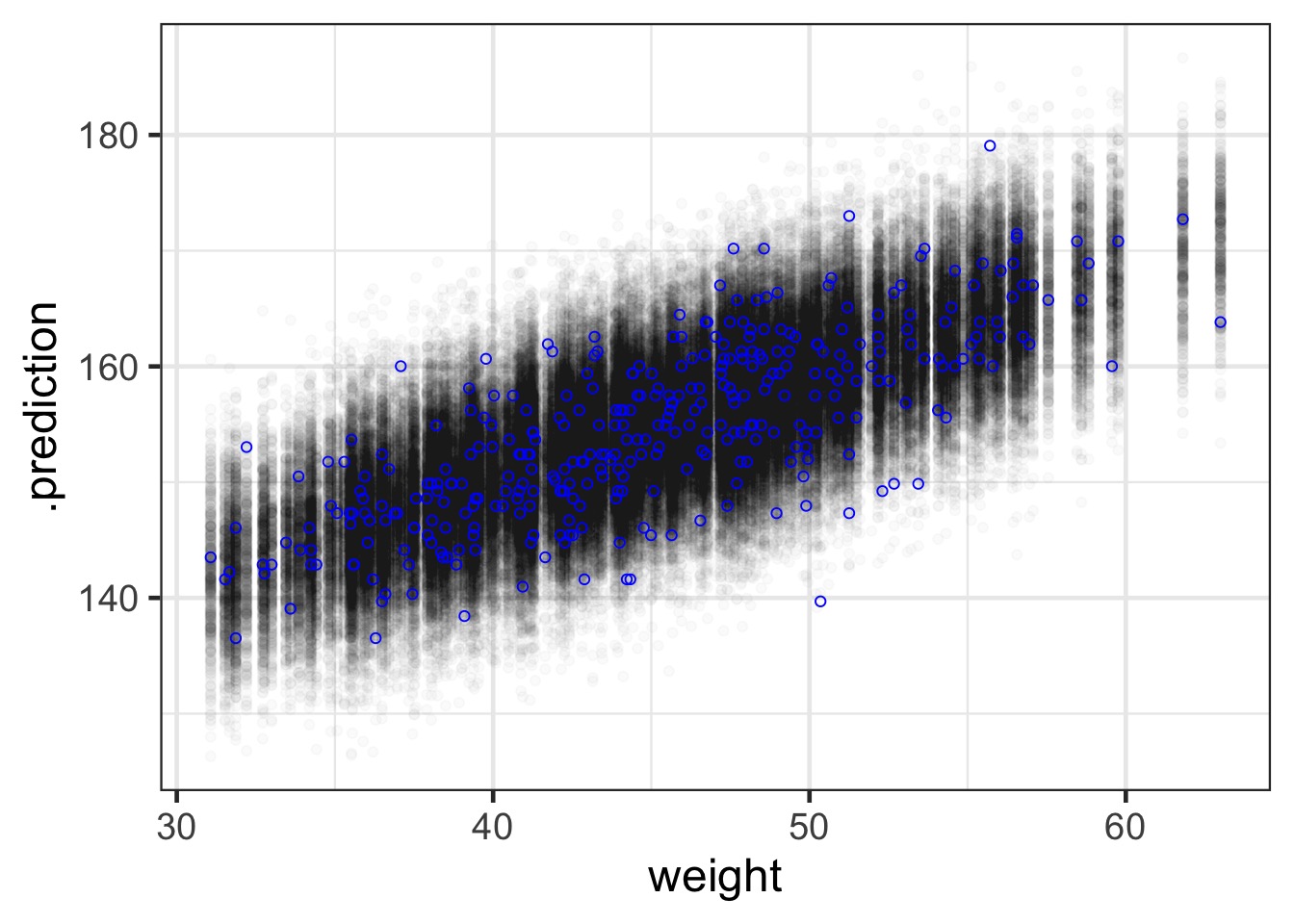
Polynomial Regression and Standardization
The Actual Data
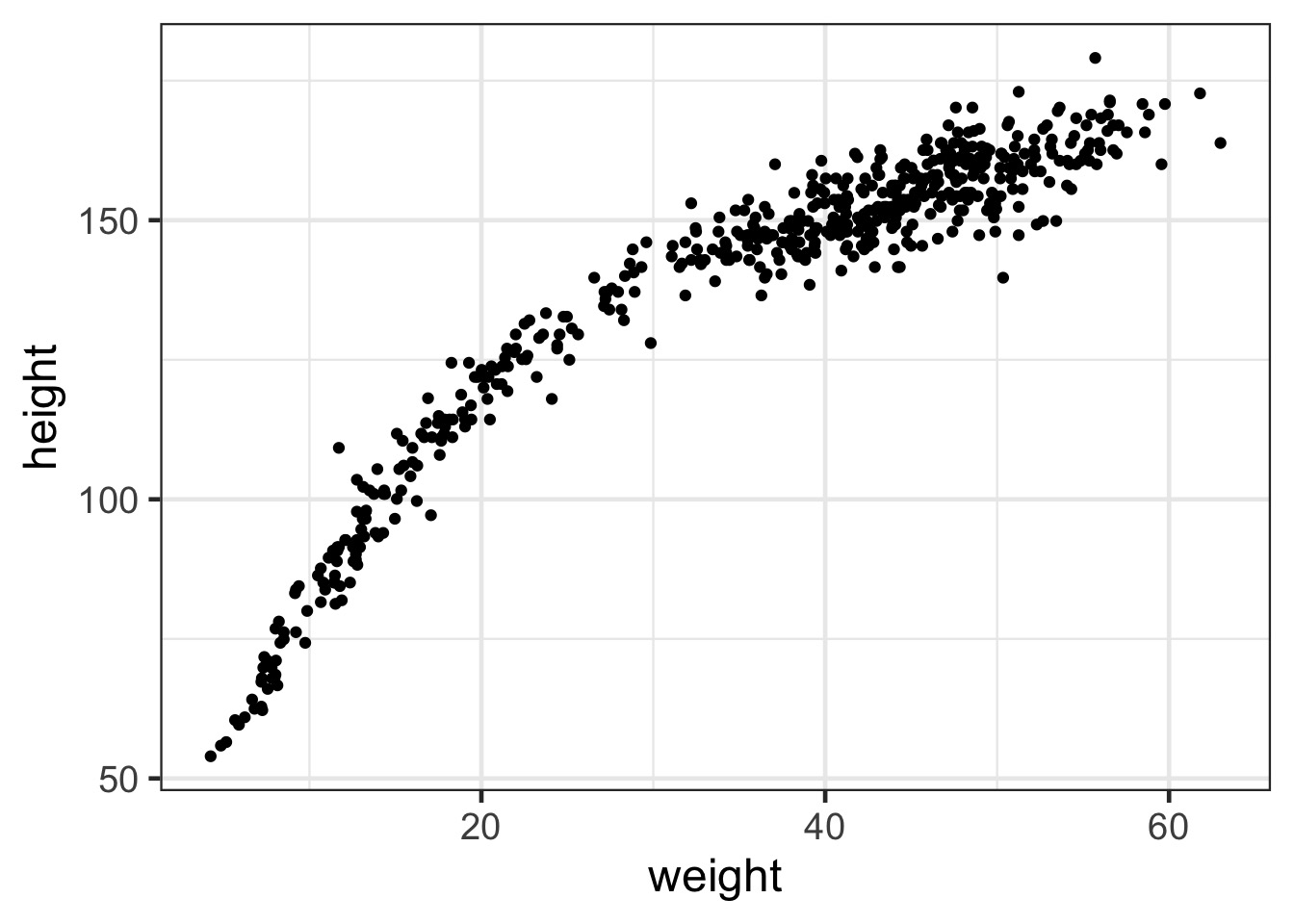
This is not linear!
Before fitting this, Standardize!
Standardizing a predictor aids in fitting
- Scale issues of different variables
Standardizing yields comparable coefficients
You don’t have to
- But if you encounter problems, it’s a good fallback
Ye Olde Z Transformation
A Nonlinear Model
Likelihood:
\(h_i \sim Normal( \mu_i, \sigma)\)
Data Generating Process
\(\mu_i = \alpha + \beta_1 x_i + \beta_2 x_i^2 + \beta_3 x_i^3\)
Prior:
\(\alpha \sim \mathcal{N}(150, 20)\)
\(\beta_j \sim \mathcal{N}(0, 10)\)
\(\sigma \sim \mathcal{U}(0,50)\)
Our Model in Code
We fit!
Before We Go Further, We CAN Look at Priors
In rethinking, we can extract prior coefficients
nonlinear_priors <- extract.prior(full_height_fit)|>
as_tibble() |>
mutate(draw = 1:n())
head(nonlinear_priors)# A tibble: 6 × 6
alpha beta1 beta2 beta3 sigma draw
<dbl[1d]> <dbl[1d]> <dbl[1d]> <dbl[1d]> <dbl[1d]> <int>
1 91.7 10.3 4.30 -0.299 46.0 1
2 269. 21.6 -2.89 15.5 25.8 2
3 153. 0.101 -0.974 5.86 14.2 3
4 168. -18.6 2.71 -9.56 43.1 4
5 183. 2.56 -2.95 -14.1 14.0 5
6 198. -12.2 -9.10 -5.33 7.90 6We Can Just Calculate Curves
Were These Good Priors?

But Did Our Model Fit?
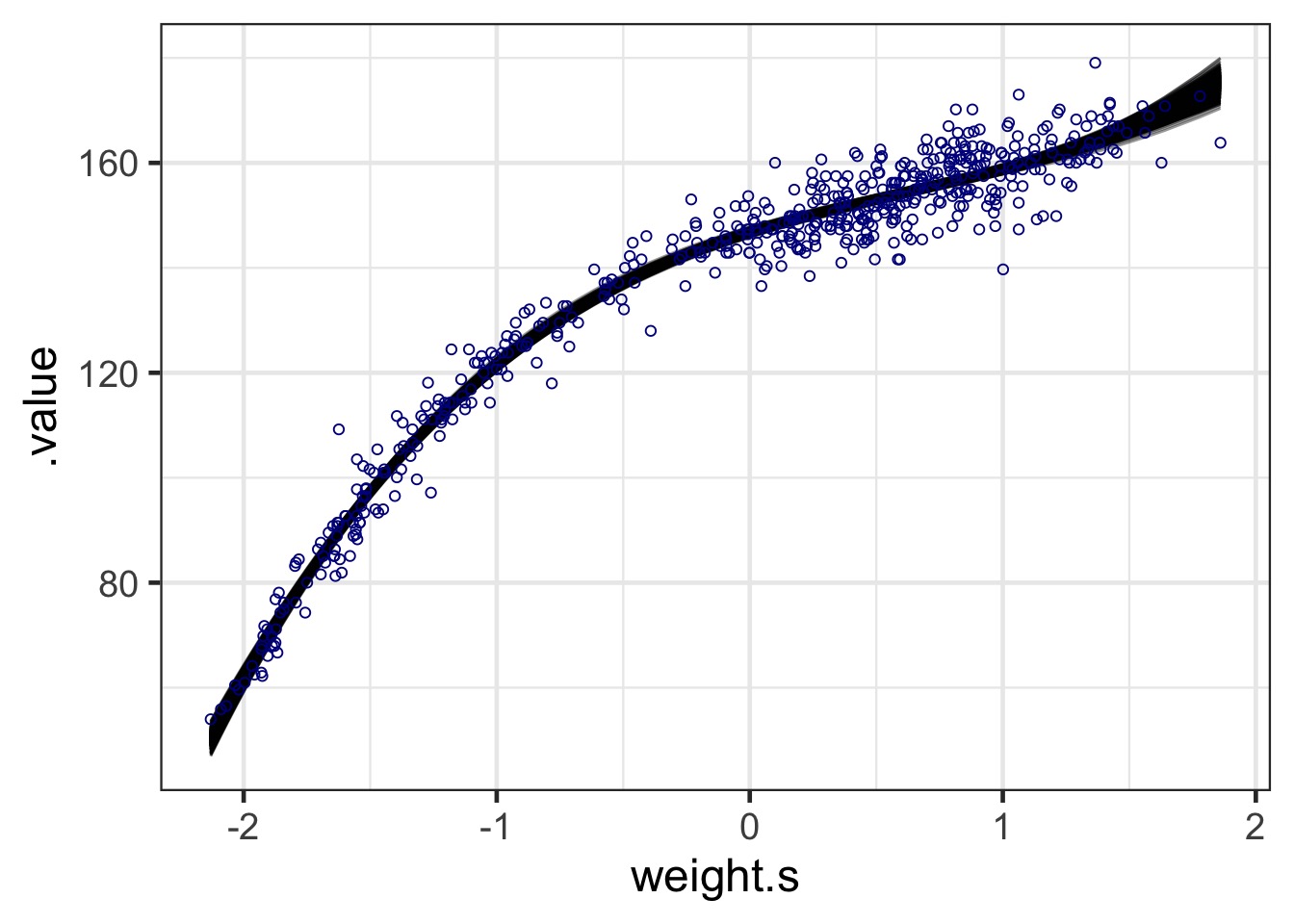
Code
poly_fits <- linpred_draws(full_height_fit, newdata = Howell1,
ndraws = 1e3)
ggplot() +
geom_line(data = poly_fits,
aes(x = weight.s, y = .value, group = .draw), alpha = 0.4) +
geom_point(data = Howell1, aes(x = weight.s, y = height),
pch = 1, color = "darkblue") #+
#scale_x_continuous(labels = round(-2:2 * sd(Howell1$weight) + mean(Howell1$weight),2))The Essence of Bayesian Modeling
Check your data and think about how geocentric you want to be.
Build a model with reasonable priors
Test your priors!
- Don’t be afraid to admit your priors are unrealistic
- If truly worried, test robustness of conclusions to prior choice
- Don’t be afraid to admit your priors are unrealistic
Evaluate the FULL implications of the posterior with samples
- This includes classic model checks
- But visualize your model fits from samples
- Do you burn down Prague?
- This includes classic model checks