Bayesian Generalized Linear Models

Our Models Until Now
Likelihood:
\(y_i \sim Normal(\mu_i, \sigma)\)
Data Generating Process
\(\mu_i = \alpha + \beta_1 x1_i + \beta_2 x2_i + ...\)
Priors:
\(\alpha \sim Normal(0, 100)\)
\(\beta_j \sim Normal(0, 100)\)
\(\sigma \sim cauchy(0,2)\)
Making the Normal General
Likelihood:
\(y_i \sim Normal(\mu_i, \sigma)\)
Data Generating Process with identity link
f(\(\mu_i) = \alpha + \beta_1 x1_i + \beta_2 x2_i + ...\)
Priors:
…
A Generalized Linear Model
Likelihood:
\(y_i \sim D(\theta_i, ...)\)
Data Generating Process with identity link
f(\(\theta_i) = \alpha + \beta_1 x1_i + \beta_2 x2_i + ...\)
Priors:
…
The Exponential Family is MaxEnt!
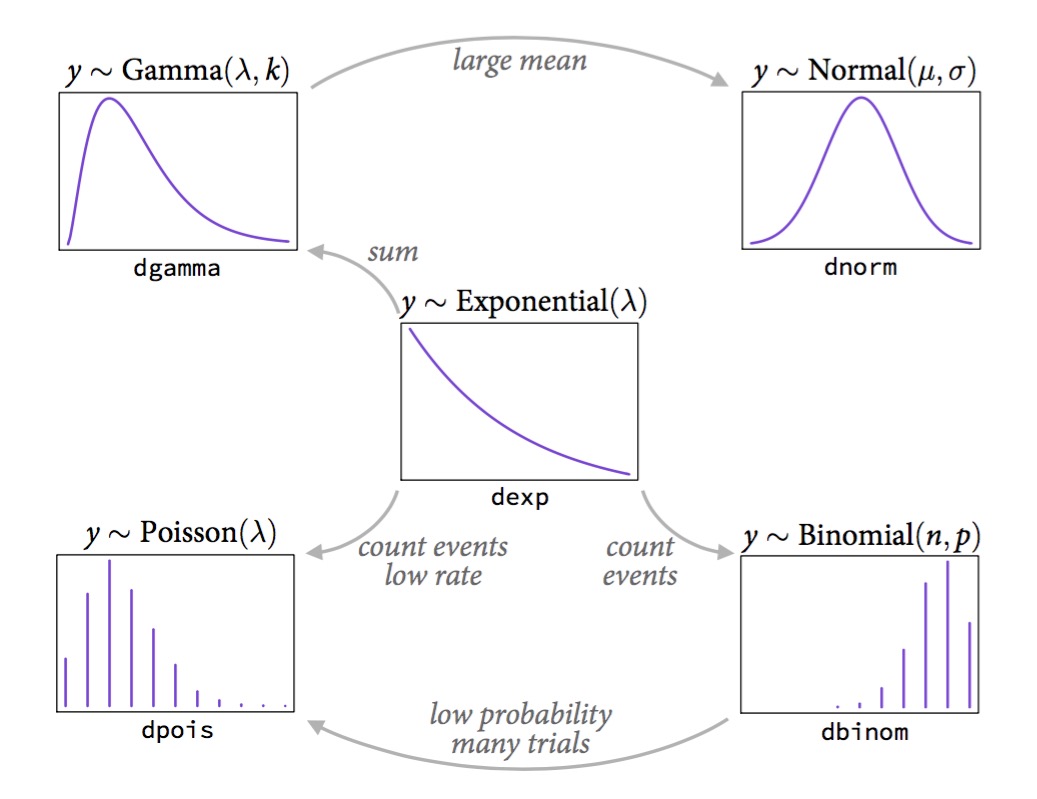
How to determine which non-normal distribution is right for you
- Use previous image to determine
- Bounded values: binomial, beta
- Counts: Poisson, multinomial, geometric
- Distances and durations: exponential, gamma (survival or event history)
- Monsters: Ranks and ordered categories
- Mixtures: Beta-binomial, gamma-Poisson, zero- inflated processes
Binomial Logistic Regression
Likelihood:
\(y_i \sim B(size, p_i)\)
Data Generating Process with identity link
logit(\(p_i) = \alpha + \beta_1 x1_i + \beta_2 x2_i + ...\)
Priors:
…
Why Binomial Logistic Regression
- Allows us to predict absolute probability of something occuring
- Allows us to determing relative change in risk due to predictors
Why a Logit Link?
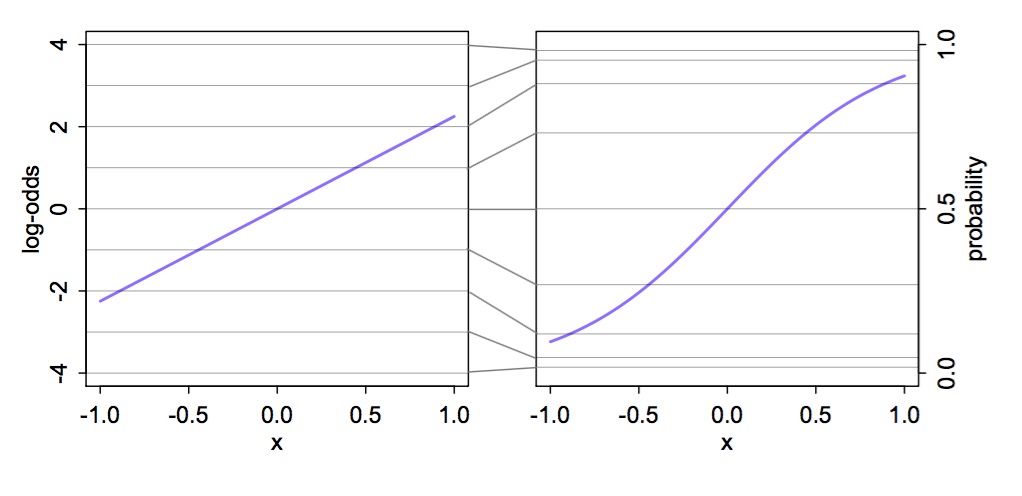
Meaning of Logit Coefficients
\[logit(p_i) = log \frac{p_i}{1-p_i} = \alpha + \beta x_i\]
- \(\frac{p_i}{1-p_i}\) is odds of something happening
- \(\beta\) is change in log odds per one unit change in \(x_i\)
- exp(\(\beta\)) is change in odds
- Change in relative risk
- exp(\(\beta\)) is change in odds
- \(p_i\) is absolute probability of something happening
- logistic(\(\alpha + \beta x_i\)) = probability
- To evaluate change in probability, choose two different \(x_i\) values
Binomial GLM in Action: Gender Discrimination in Graduate Admissions

Our data: Berkeley
dept applicant.gender admit reject applications
1 A male 512 313 825
2 A female 89 19 108
3 B male 353 207 560
4 B female 17 8 25
5 C male 120 205 325
6 C female 202 391 593What model would you build?
dept applicant.gender admit reject applications
1 A male 512 313 825
2 A female 89 19 108
3 B male 353 207 560
4 B female 17 8 25
5 C male 120 205 325
6 C female 202 391 593One Model
What do Priors Imply in a GLMS?
Visualize what that means!
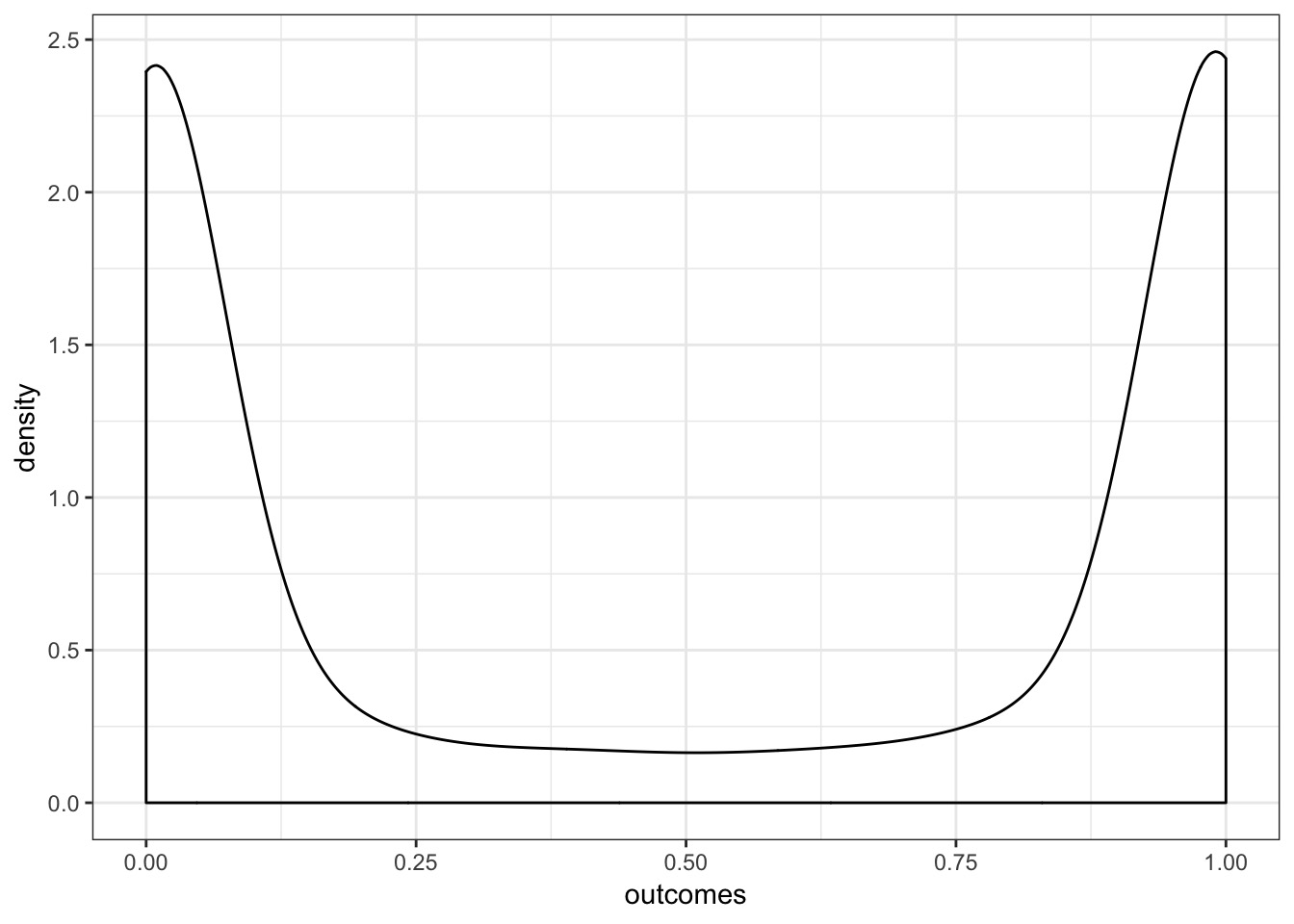
A Flat Prior in Logit Space Might Not Be Flat!

Let’s try a few SDs to see what works!
What is our flat prior?
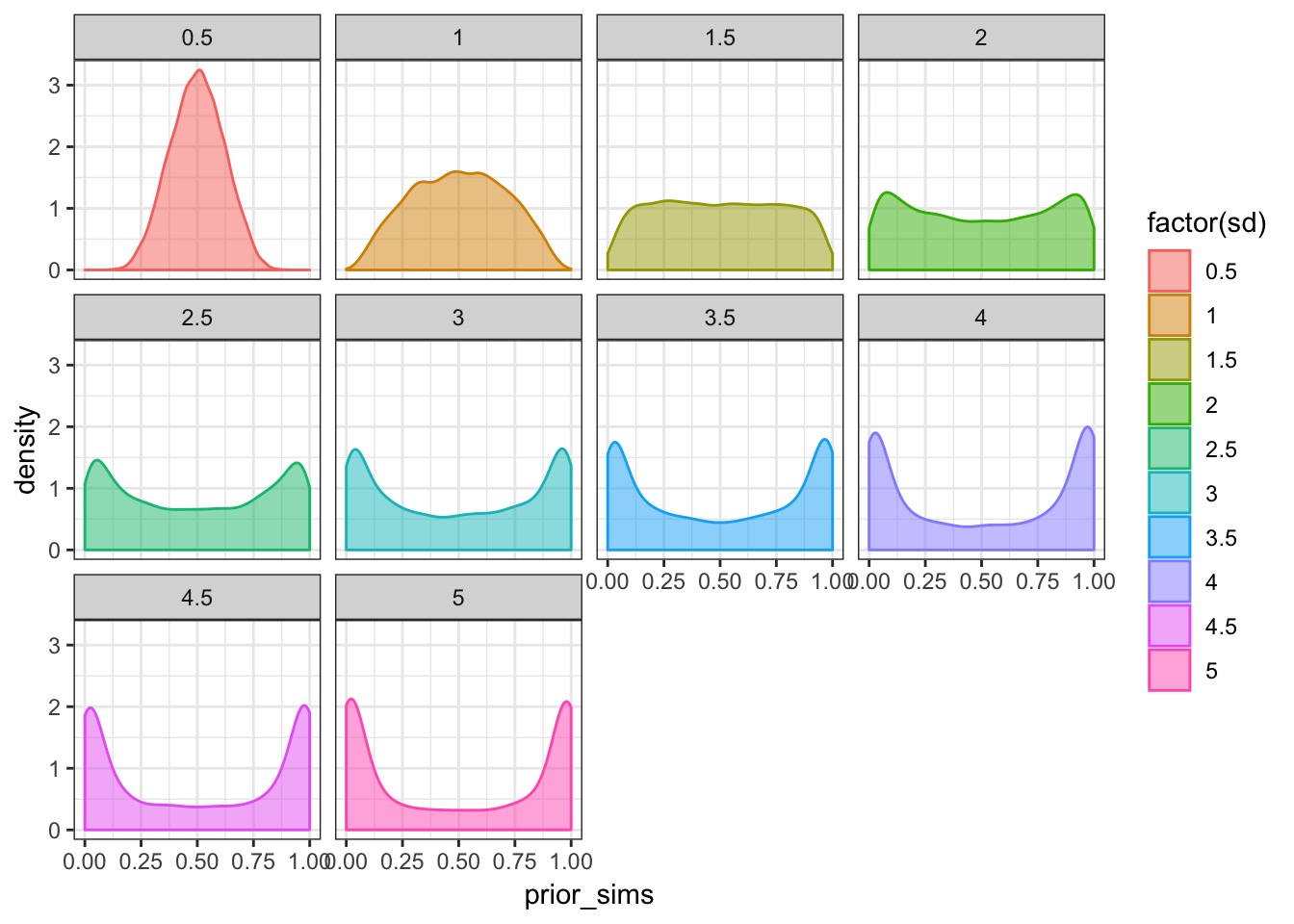
A New Model
Fit the Model!
Results… men do better!
Mean StdDev 5.5% 94.5%
a[1] -0.83 0.05 -0.91 -0.75
a[2] -0.22 0.04 -0.28 -0.16What it means
[1] 0.3036451[1] 0.4452208Relative comparison: Male Advantage
[1] 1.840247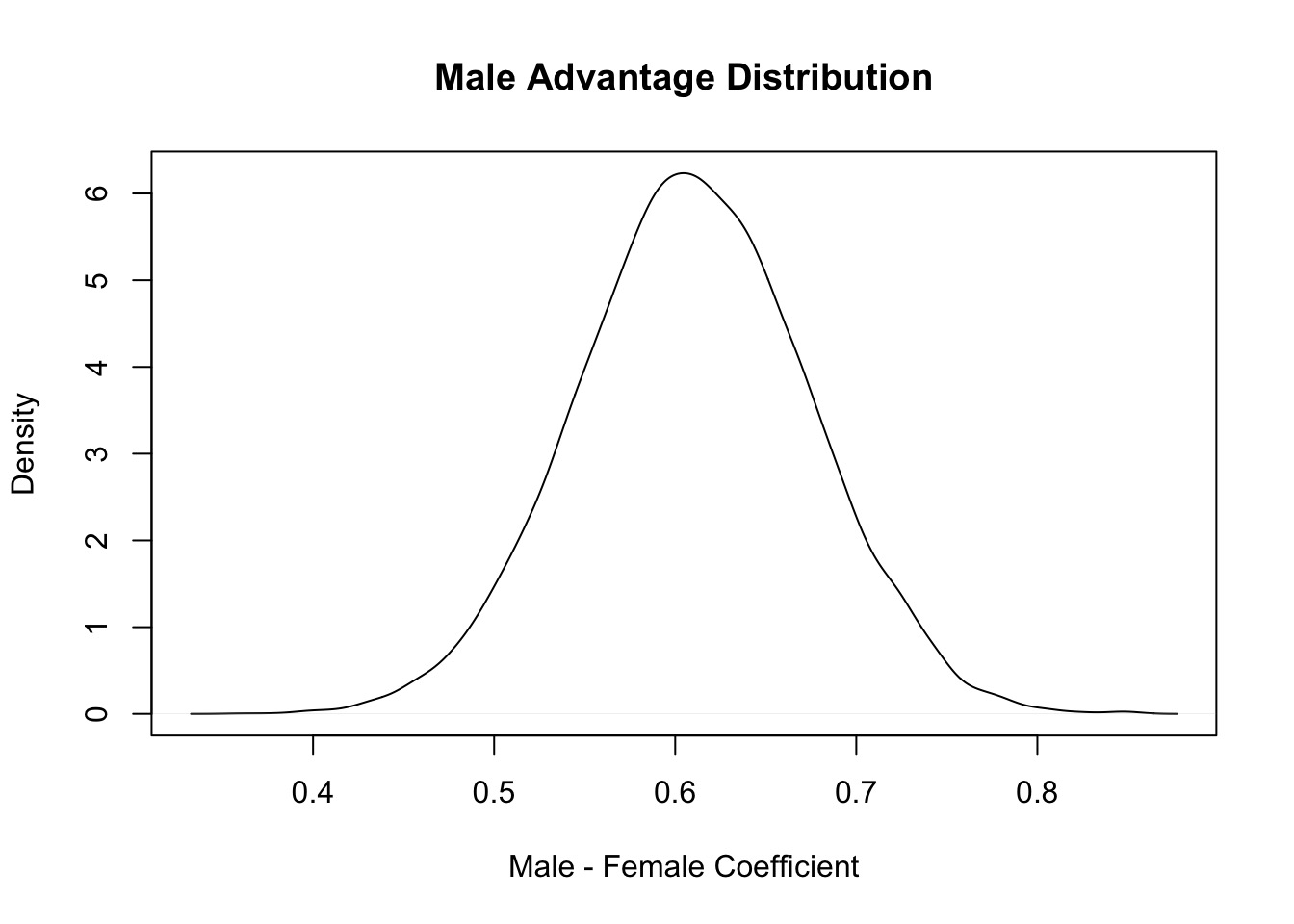
Residuals and Fits
post_pred <- sim(fit_gender)
post_fits <- apply(post_pred, 2, median)
post_res <- sapply(1:nrow(UCBadmit),
function(i) UCBadmit$admit[i] - post_pred[,i])
res_frame <- tibble(fitted = post_fits,
res = apply(post_res, 2, median),
lwr_res = res - apply(post_res, 2, sd),
upr_res = res + apply(post_res, 2, sd))But - can’t use raw residuals
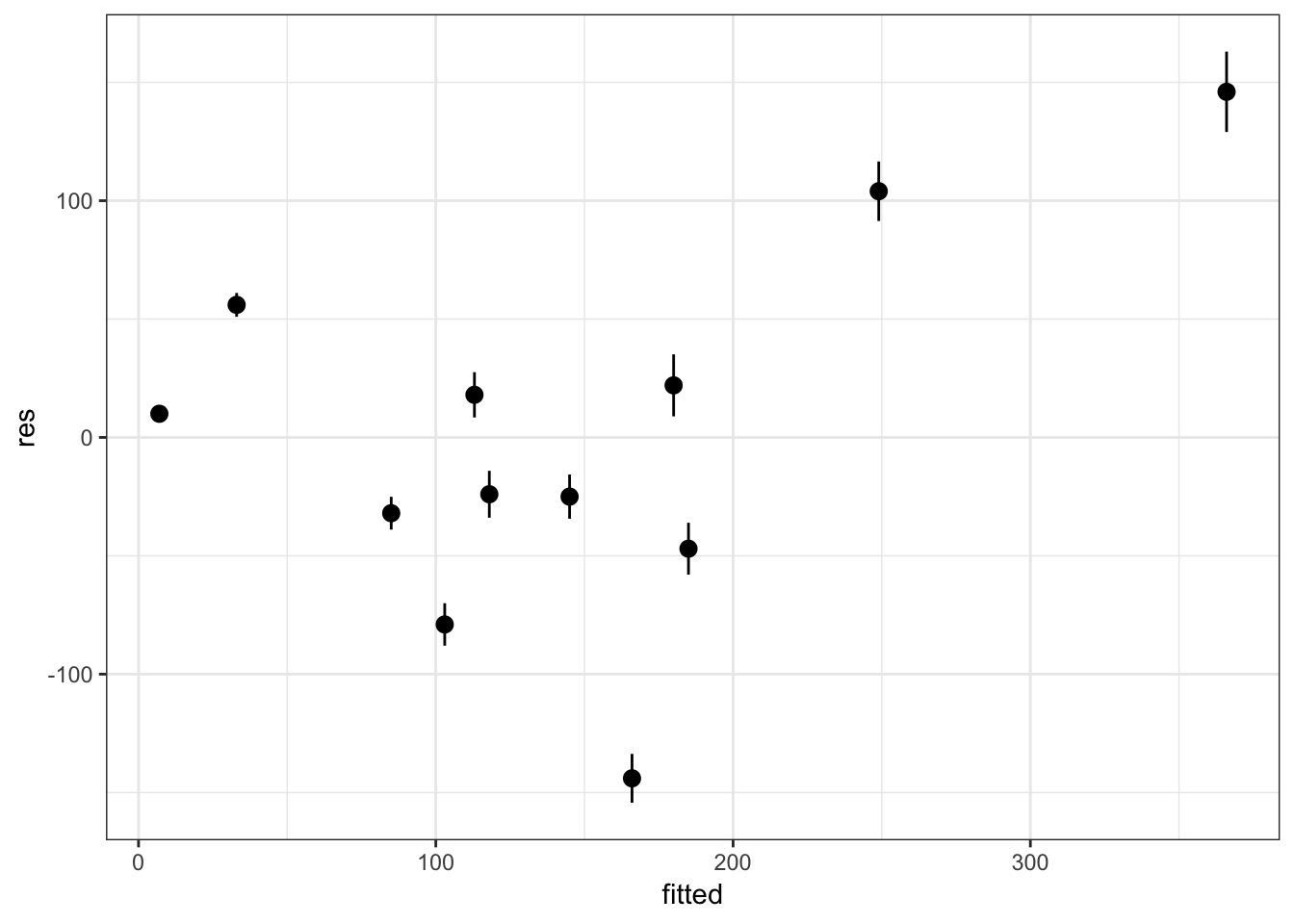
But - can’t use raw residuals
library(tidyr)
res_tib <- as_tibble(post_res) %>%
gather(variable, value) %>%
group_by(variable) %>%
summarize(res = median(value))
ggplot(res_tib, aes(sample = res)) +
geom_qq() + geom_qq_line()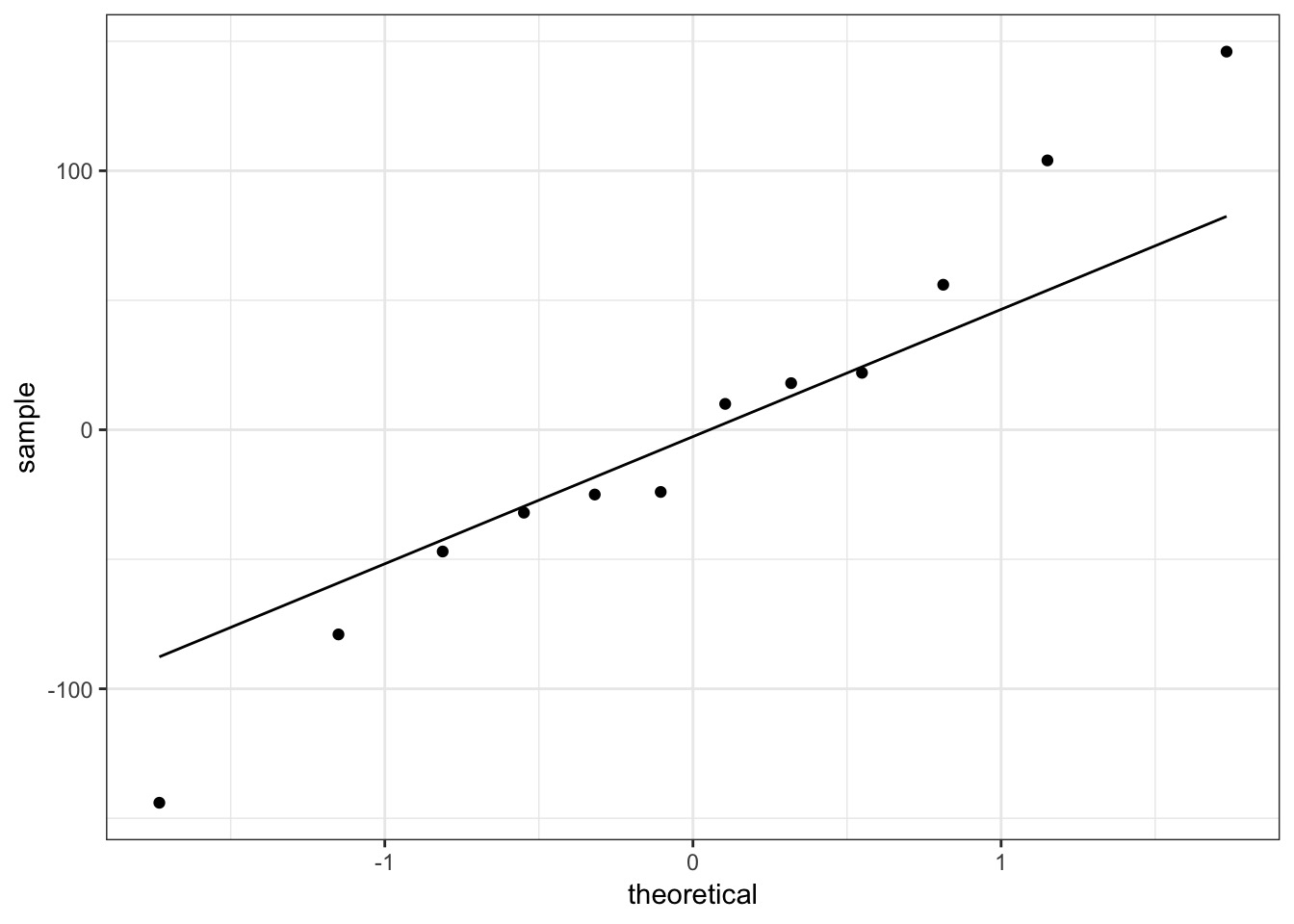
Quantiles of Residuals via Simulation
- Remember that QQ Norm plots don’t work?
- But we can get quantiles of predictions relative to posterior prediction
- What % are < the observed value
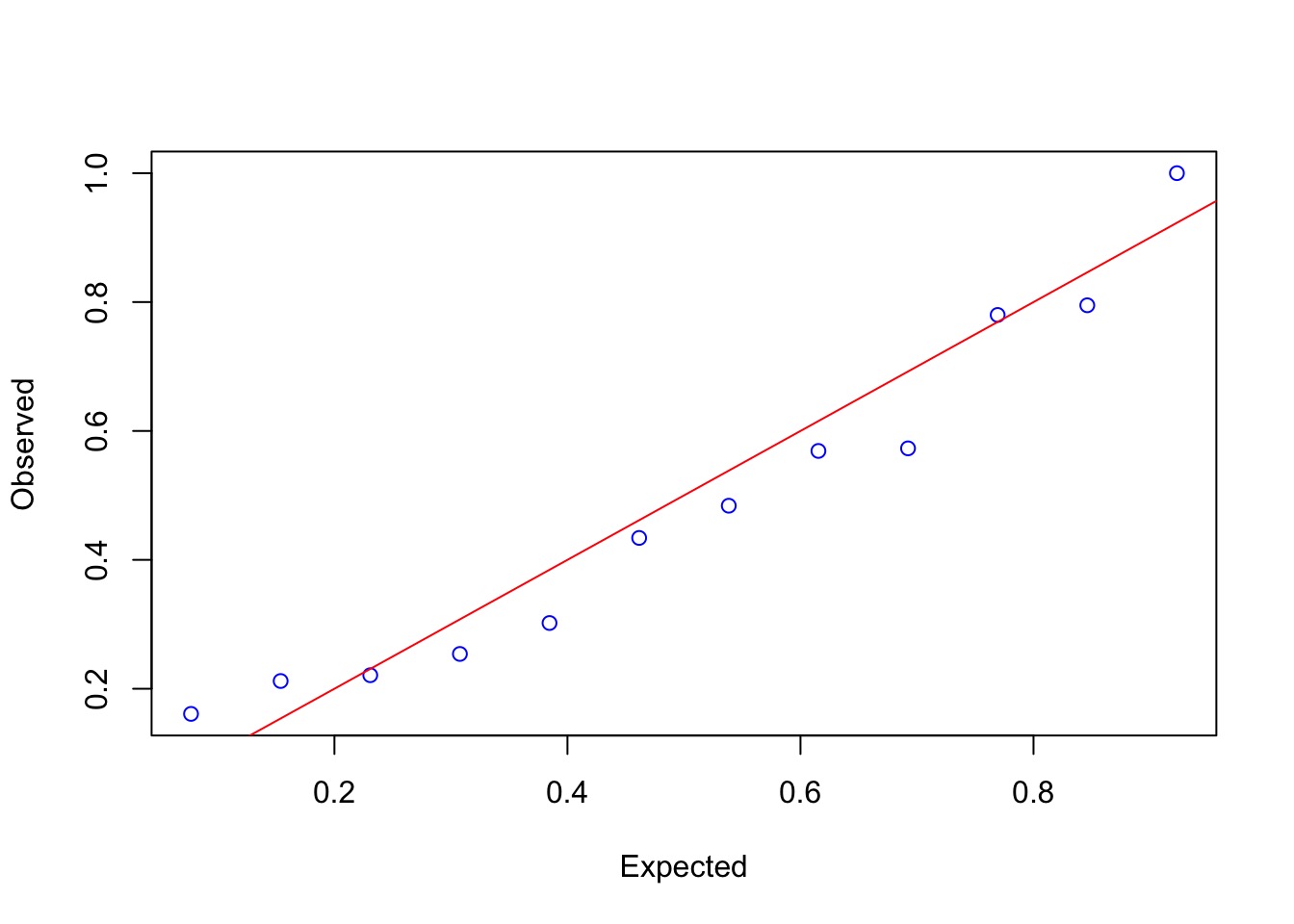
QQ Unif Looks..not great….
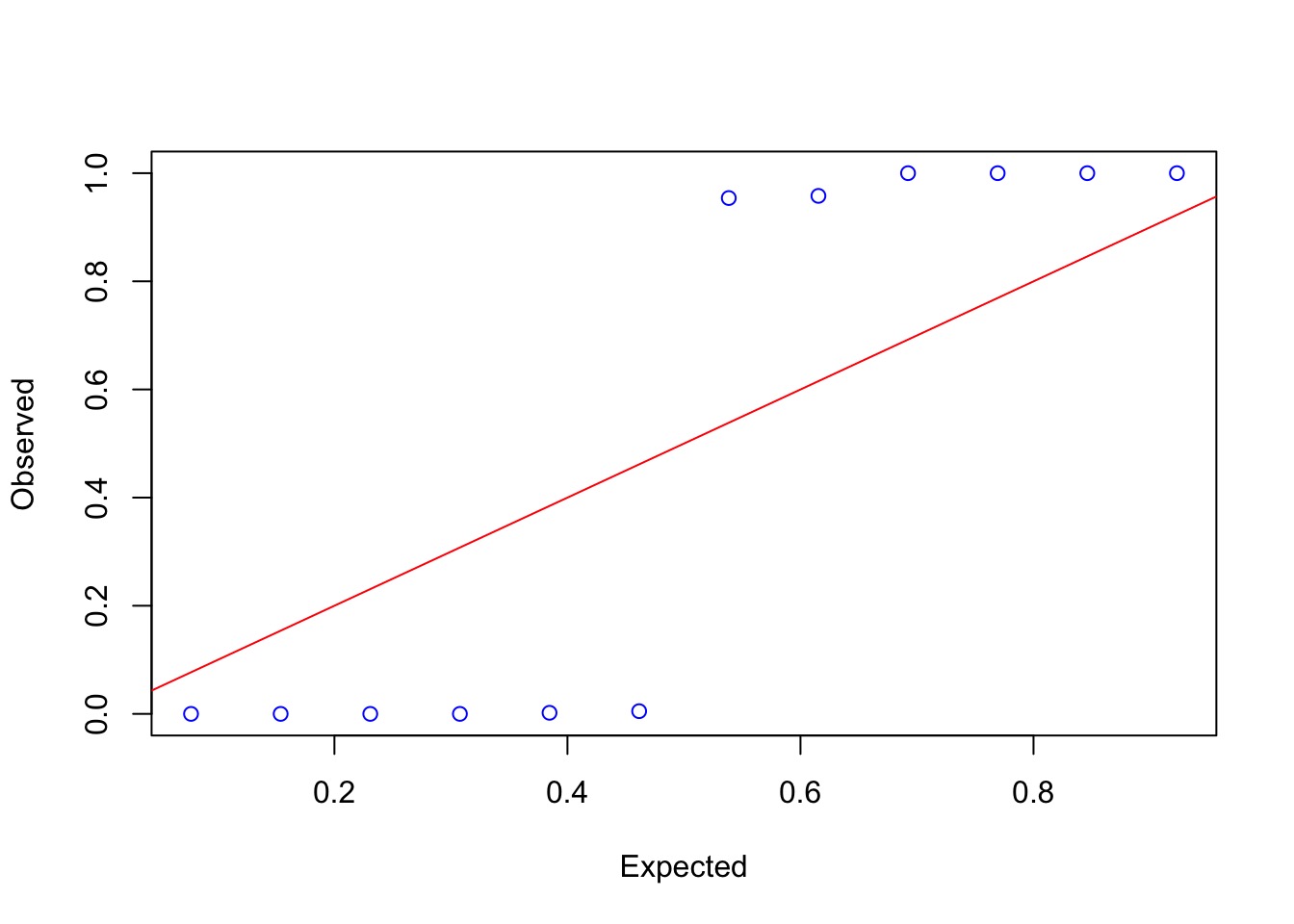
Why wasn’t this a good model?
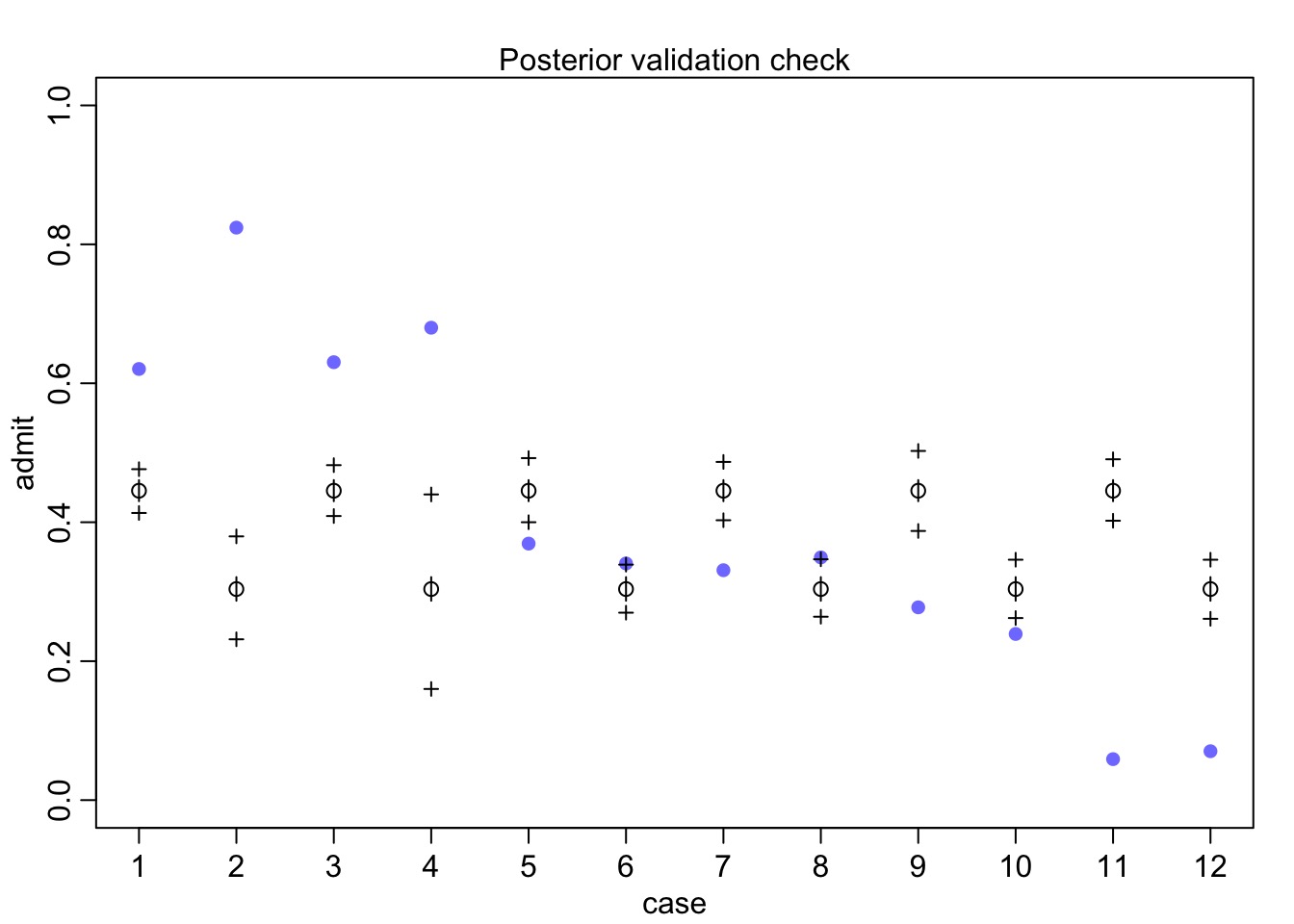
Why wasn’t this a good model?
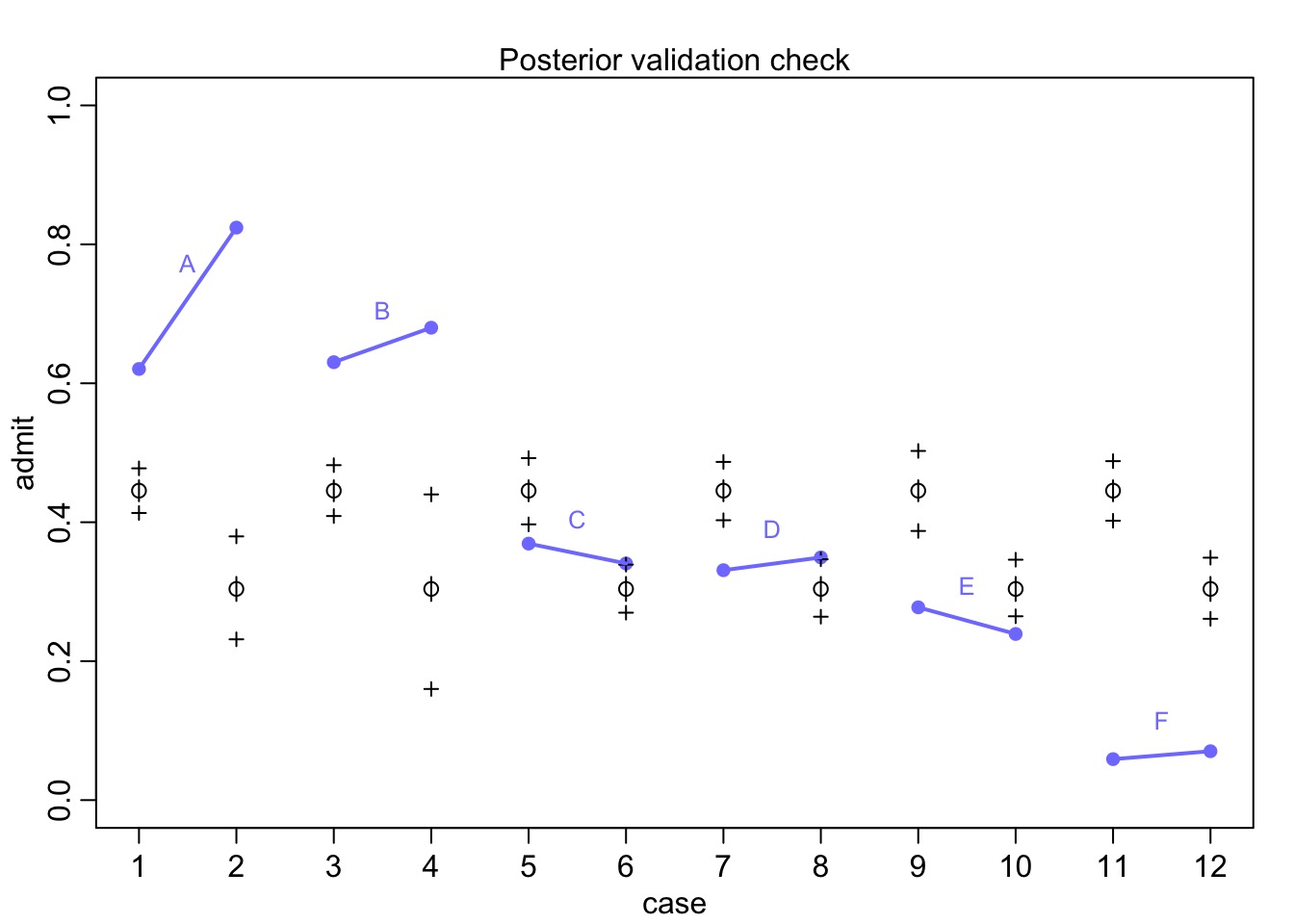
Departments Not Centered on 0 when we consider Department!
UCB_full <- cbind(UCBadmit, res_frame)
ggplot(UCB_full, mapping = aes(x = dept, y = res)) +
geom_boxplot()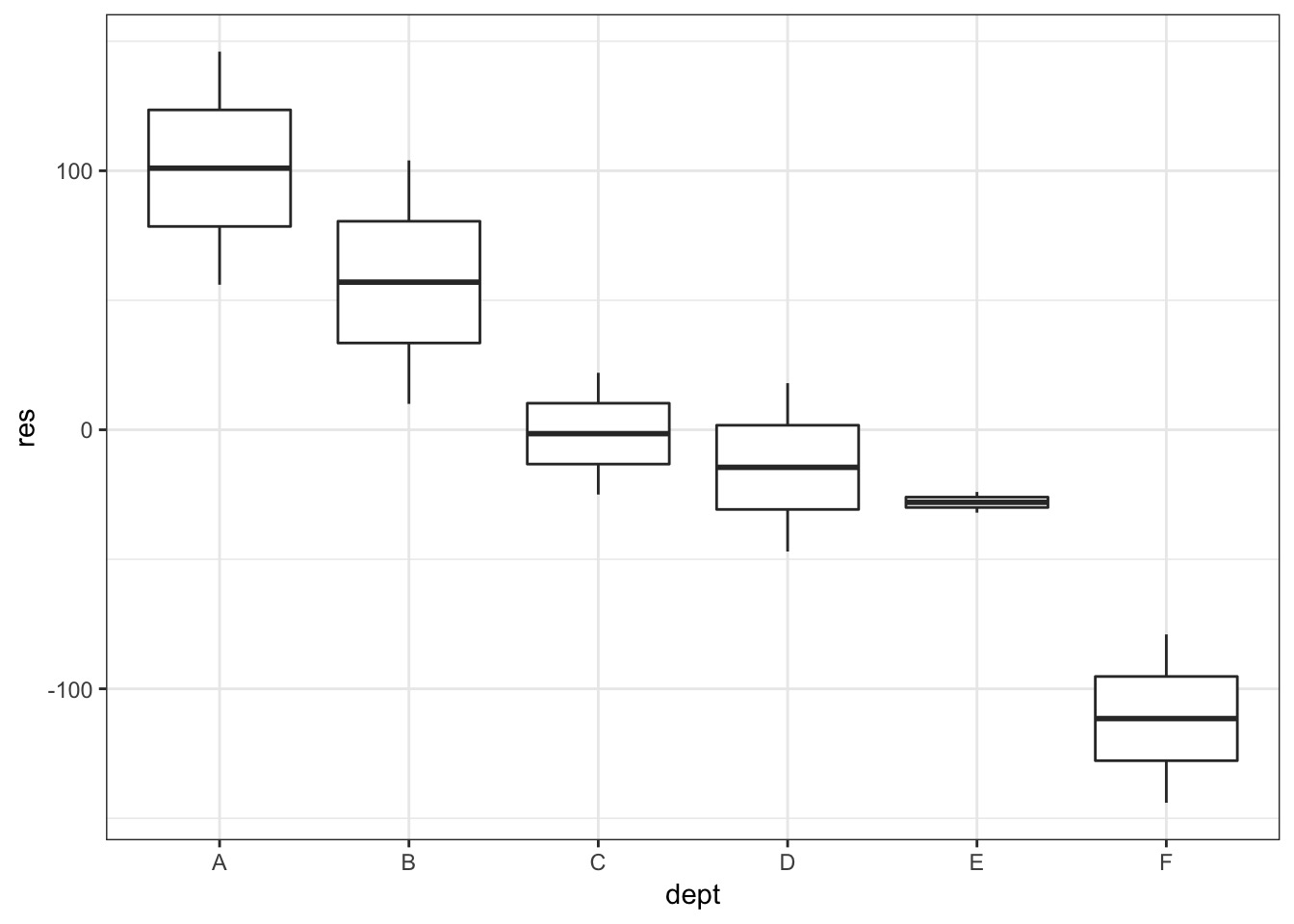
So, department…
#Make a dept index
UCBadmit$dept_id <- as.numeric(UCBadmit$dept)
mod_gender_dept <- alist(
#likelihood
admit ~ dbinom(applications, p),
#Data generating process
logit(p) <- a[dept_id] + b[gender],
#priors
a[dept_id] ~ dnorm(0,0.5),
b[gender] ~ dnorm(0,0.5)
)
fit_gender_dept <- map(mod_gender_dept, UCBadmit)Did it work?
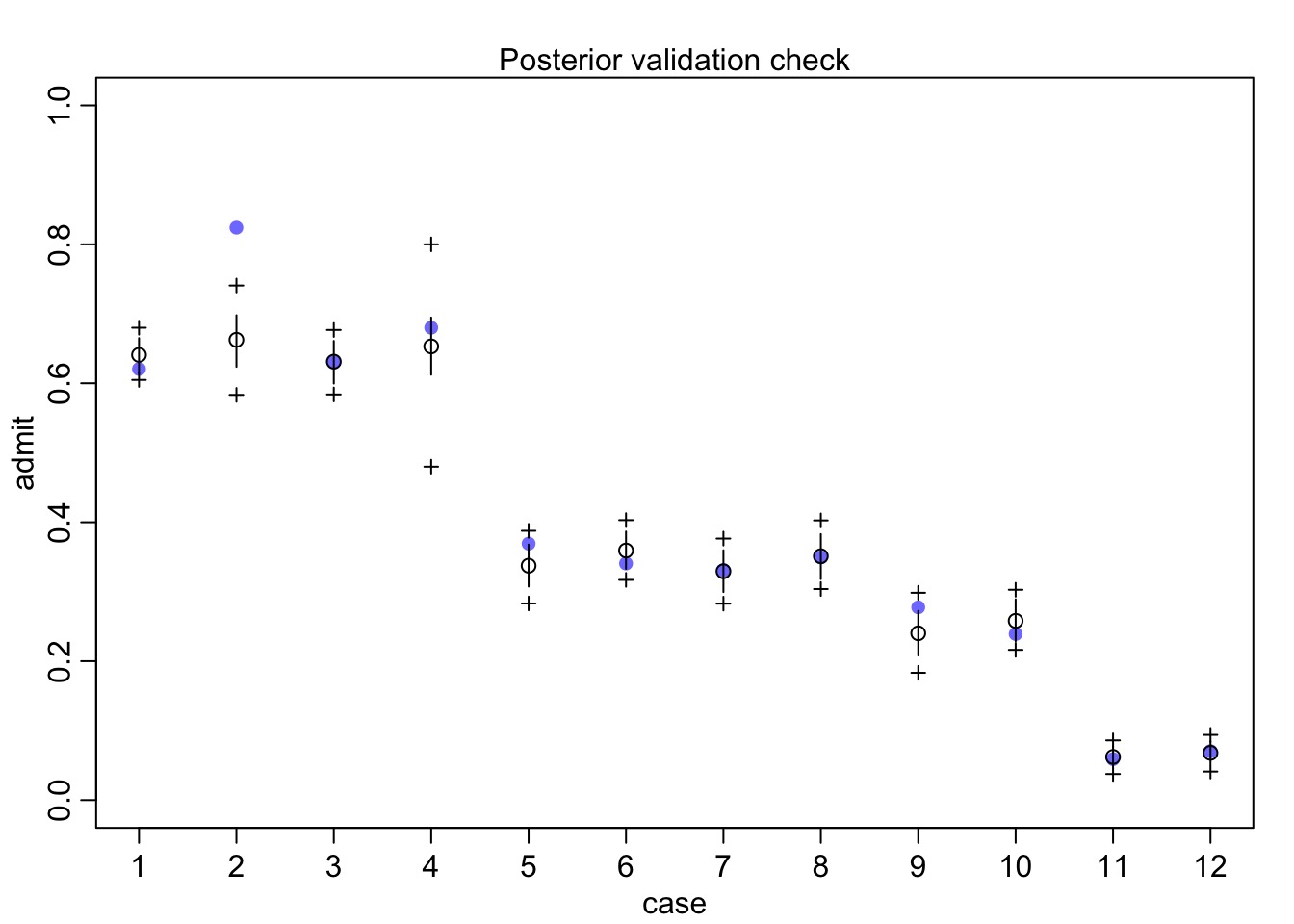
QQ Unif…
#sim gets predictions
post_pred_2 <- sim(fit_gender_dept, refresh=0)
#figure out how much of each column < observation
quants_2 <- sapply(1:nrow(UCBadmit), function(i){
#what fraction of predictions < the observation
sum(post_pred_2[,i] < UCBadmit$admit[i])/nrow(post_pred_2)
})
gap::qqunif(quants_2, logscale=FALSE)Quantile Residuals say Yes!
Outcomes
Mean StdDev 5.5% 94.5%
a[1] 1.07 0.19 0.76 1.37
a[2] 1.01 0.20 0.70 1.32
a[3] -0.16 0.19 -0.46 0.14
a[4] -0.19 0.19 -0.50 0.11
a[5] -0.62 0.20 -0.93 -0.30
a[6] -2.04 0.21 -2.38 -1.69
b[1] -0.43 0.18 -0.72 -0.13
b[2] -0.50 0.18 -0.79 -0.21Tool Use and Poisson Regression
What drives tool use?
culture population contact total_tools mean_TU
1 Malekula 1100 low 13 3.2
2 Tikopia 1500 low 22 4.7
3 Santa Cruz 3600 low 24 4.0
4 Yap 4791 high 43 5.0
5 Lau Fiji 7400 high 33 5.0
6 Trobriand 8000 high 19 4.0How to Handle the data?
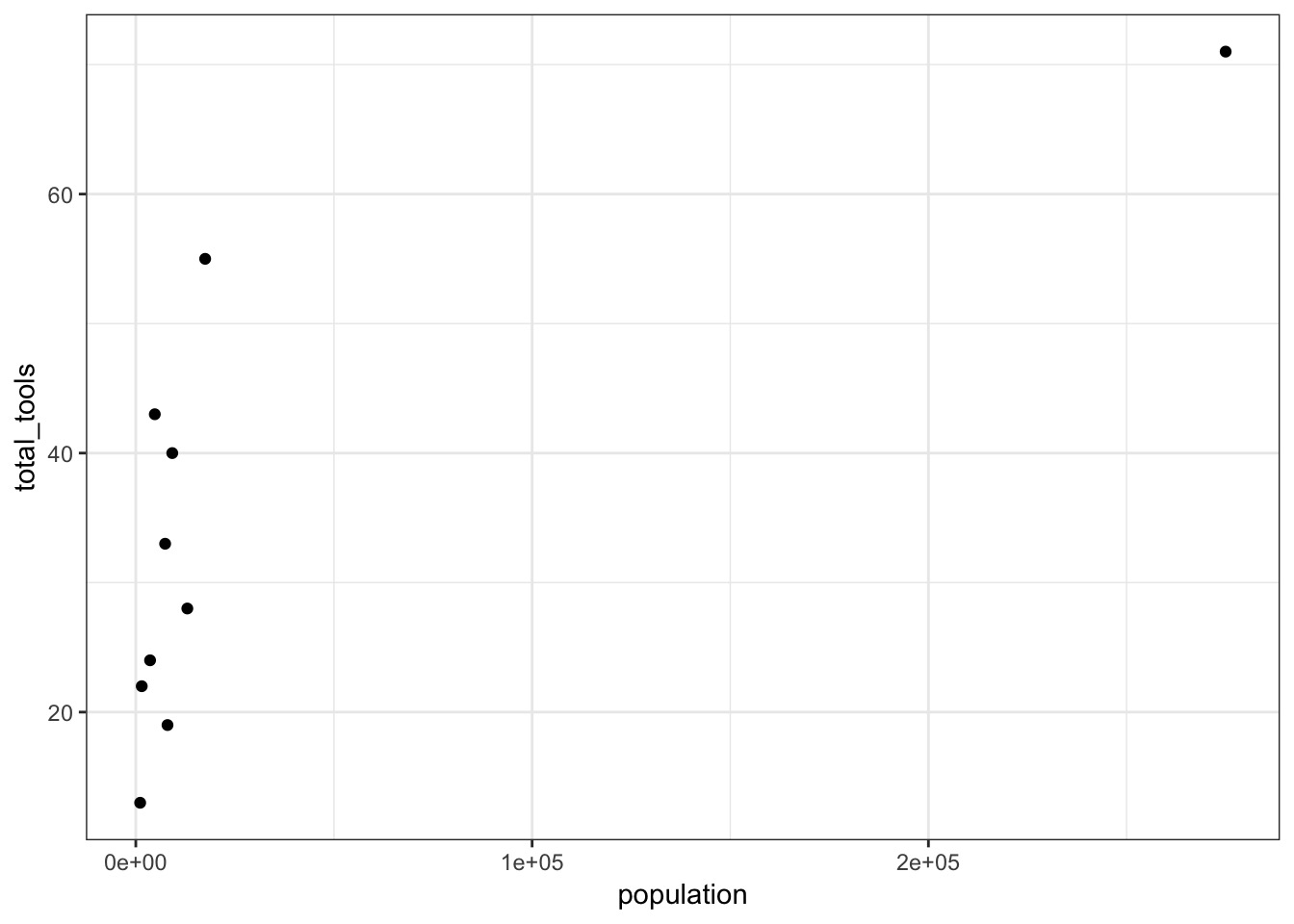
OK, Population scale is off - log it?
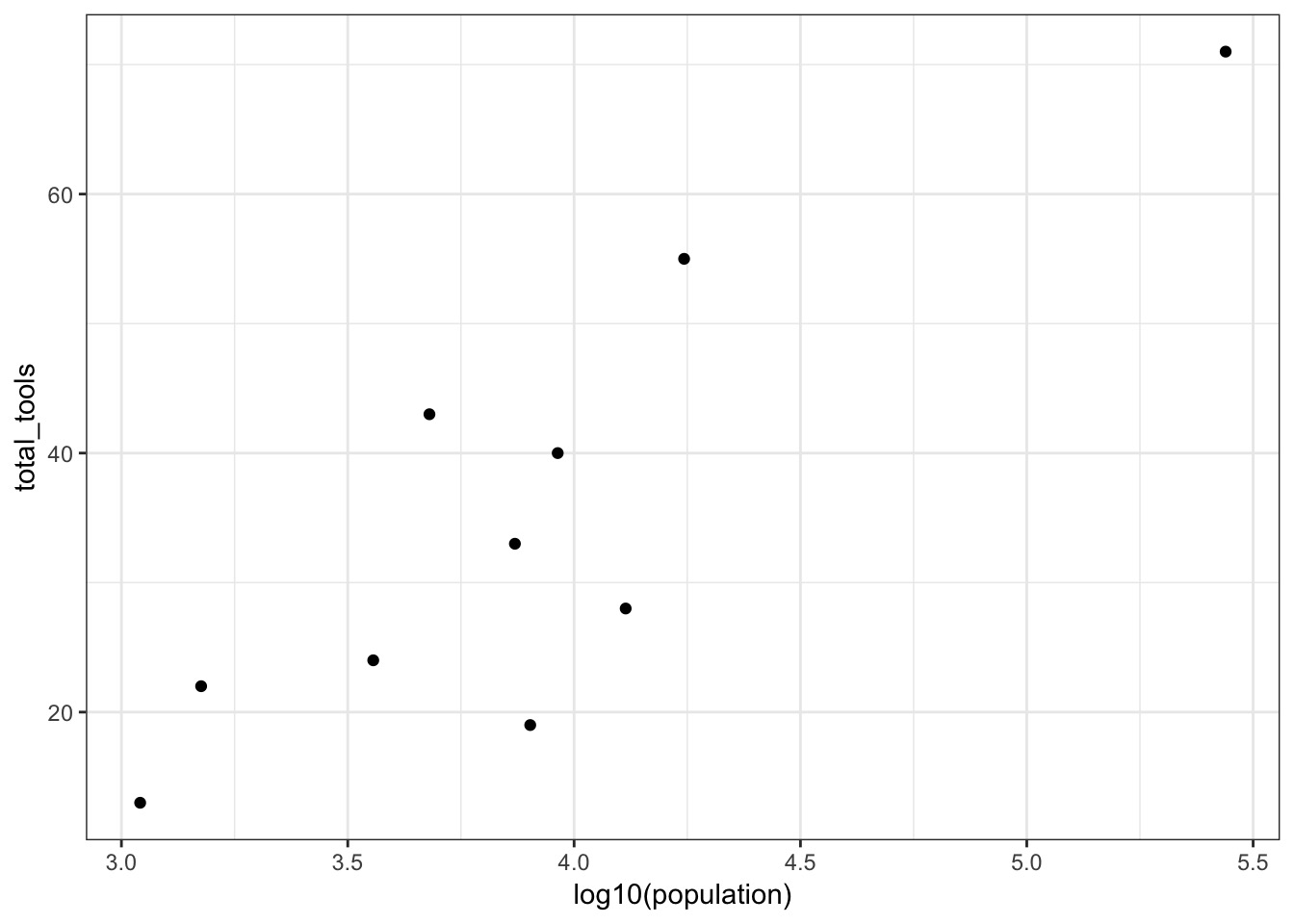
Center Population, as our smallest still have 1K People!
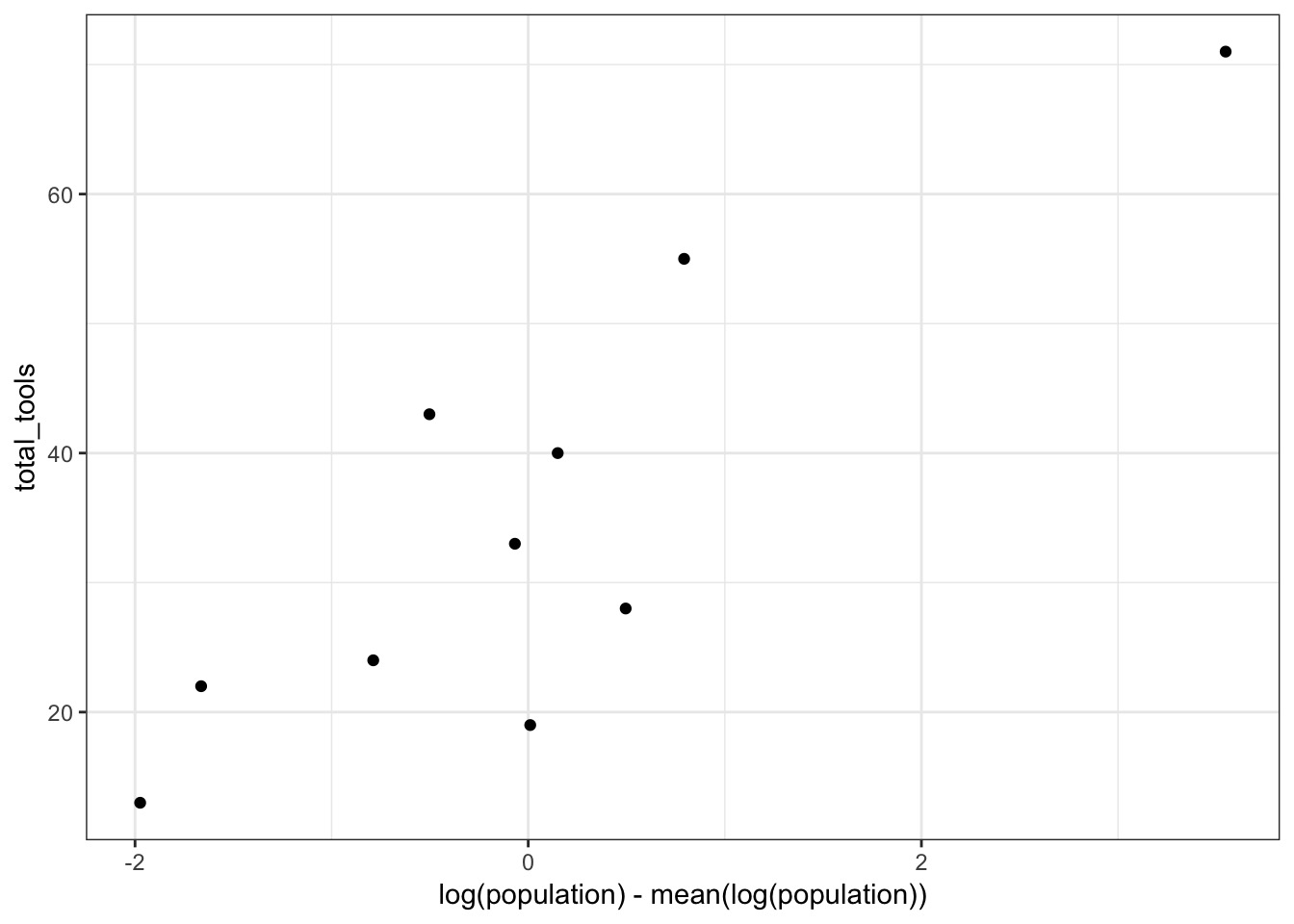
A Simple Model
What should our priors be? Data

Try out a few
Visualize Priors (data centered on ~40)
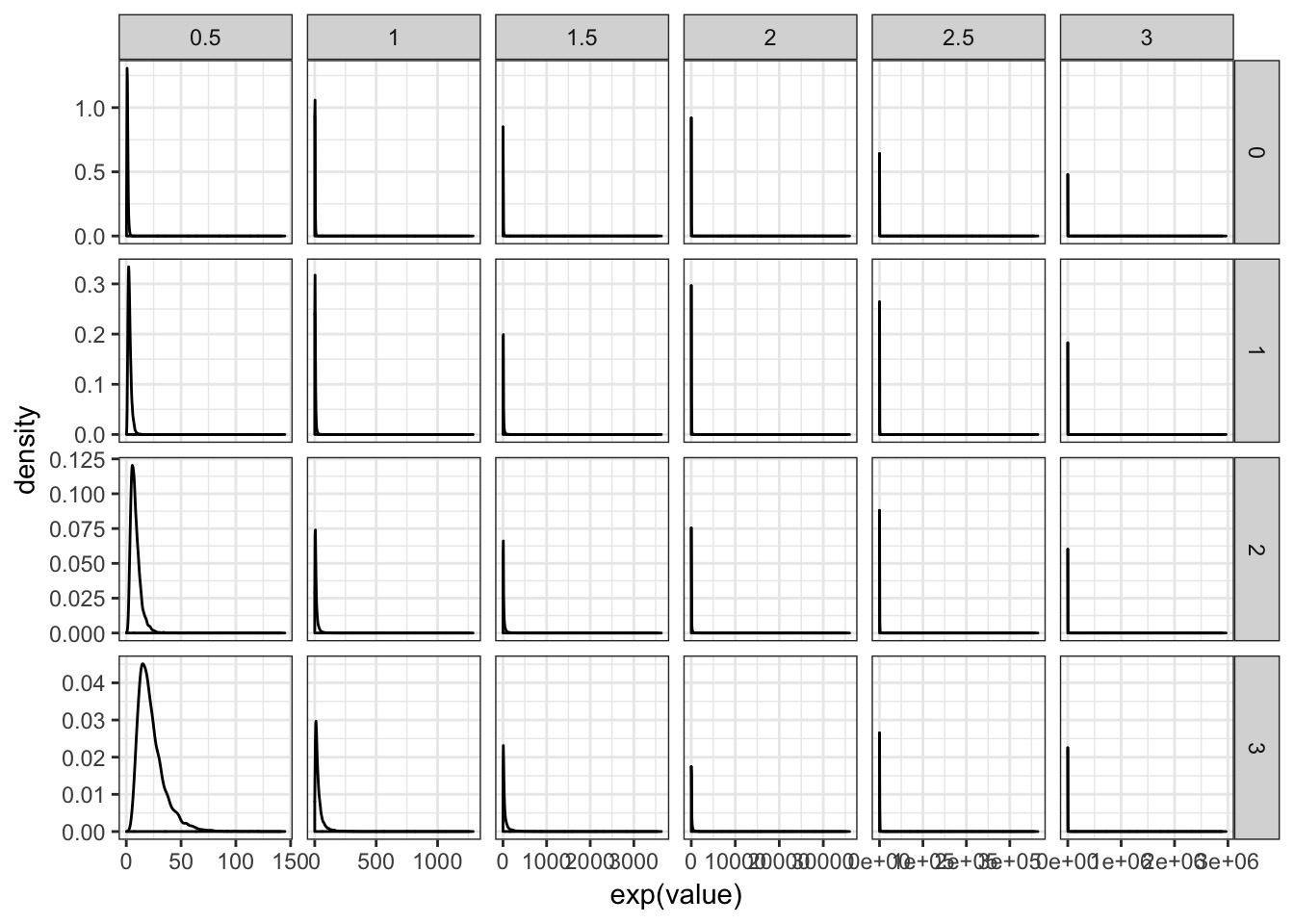
What about b?
What about b?
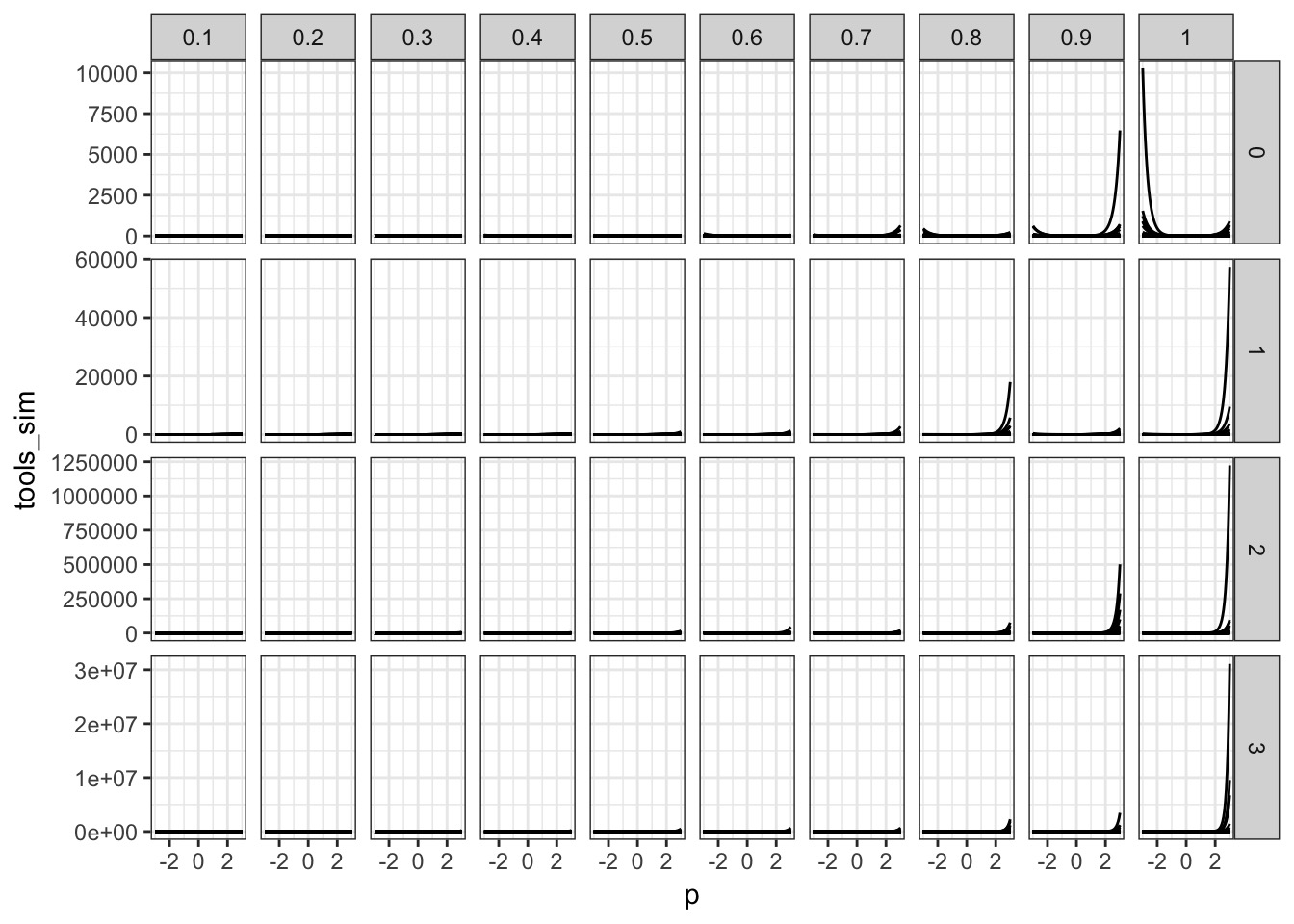
What about b?
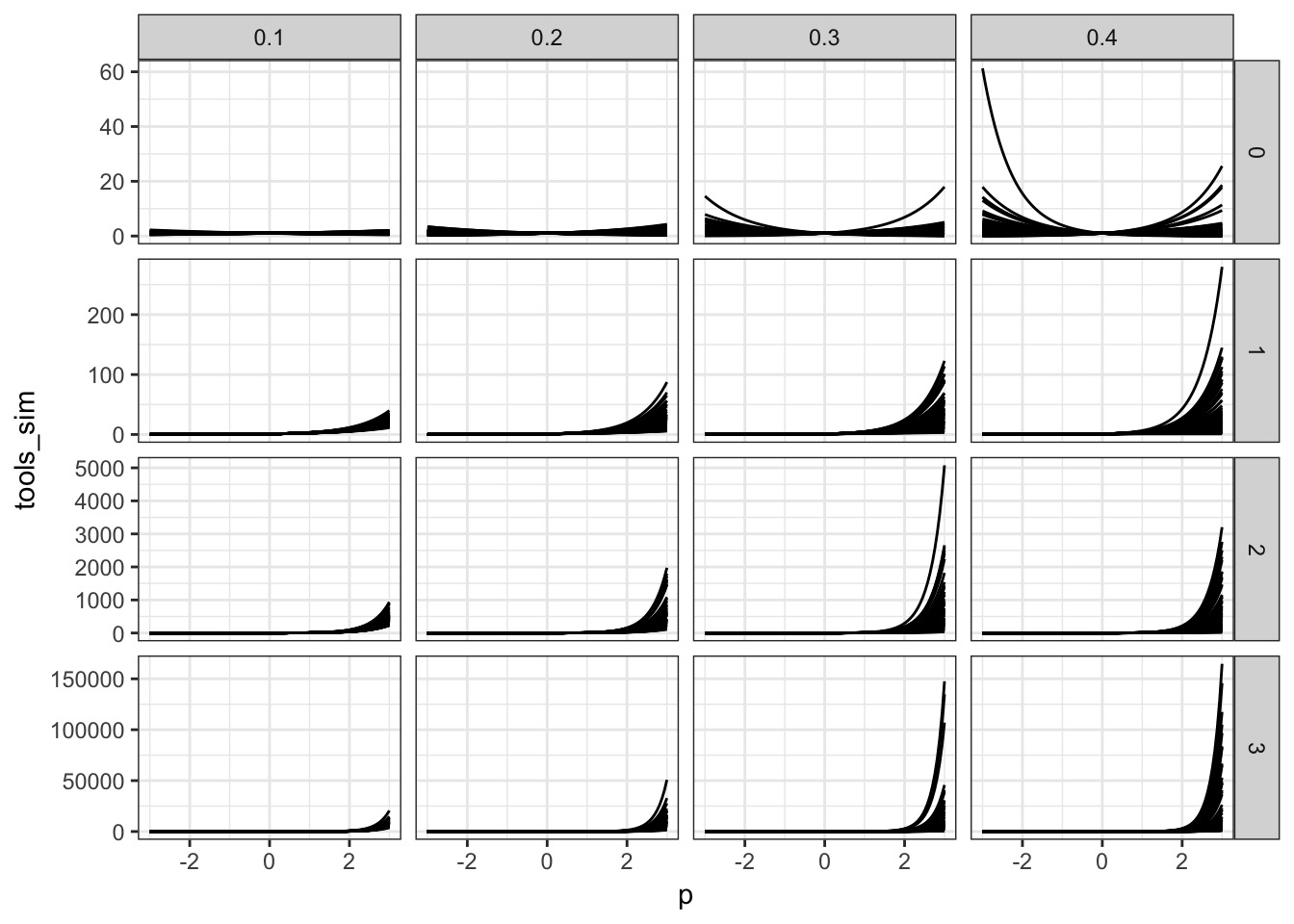
A Simple Model
Kline$P <- log(Kline$population) - mean(log(Kline$population))
kline_mod <- alist(
#likelihood
total_tools ~ dpois(lambda),
#DGP
log(lambda) <- a + b * P,
#priors
a ~ dnorm(3,0.5),
b ~ dnorm(1,0.2)
)
kline_fit <- map2stan(kline_mod, data = Kline)
SAMPLING FOR MODEL 'total_tools ~ dpois(lambda)' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 1.4e-05 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.14 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 0.017101 seconds (Warm-up)
Chain 1: 0.017591 seconds (Sampling)
Chain 1: 0.034692 seconds (Total)
Chain 1:
SAMPLING FOR MODEL 'total_tools ~ dpois(lambda)' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 6e-06 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.06 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: WARNING: No variance estimation is
Chain 1: performed for num_warmup < 20
Chain 1:
Chain 1: Iteration: 1 / 1 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 2e-06 seconds (Warm-up)
Chain 1: 1.9e-05 seconds (Sampling)
Chain 1: 2.1e-05 seconds (Total)
Chain 1:
[ 100 / 1000 ]
[ 200 / 1000 ]
[ 300 / 1000 ]
[ 400 / 1000 ]
[ 500 / 1000 ]
[ 600 / 1000 ]
[ 700 / 1000 ]
[ 800 / 1000 ]
[ 900 / 1000 ]
[ 1000 / 1000 ]##Did we converge?
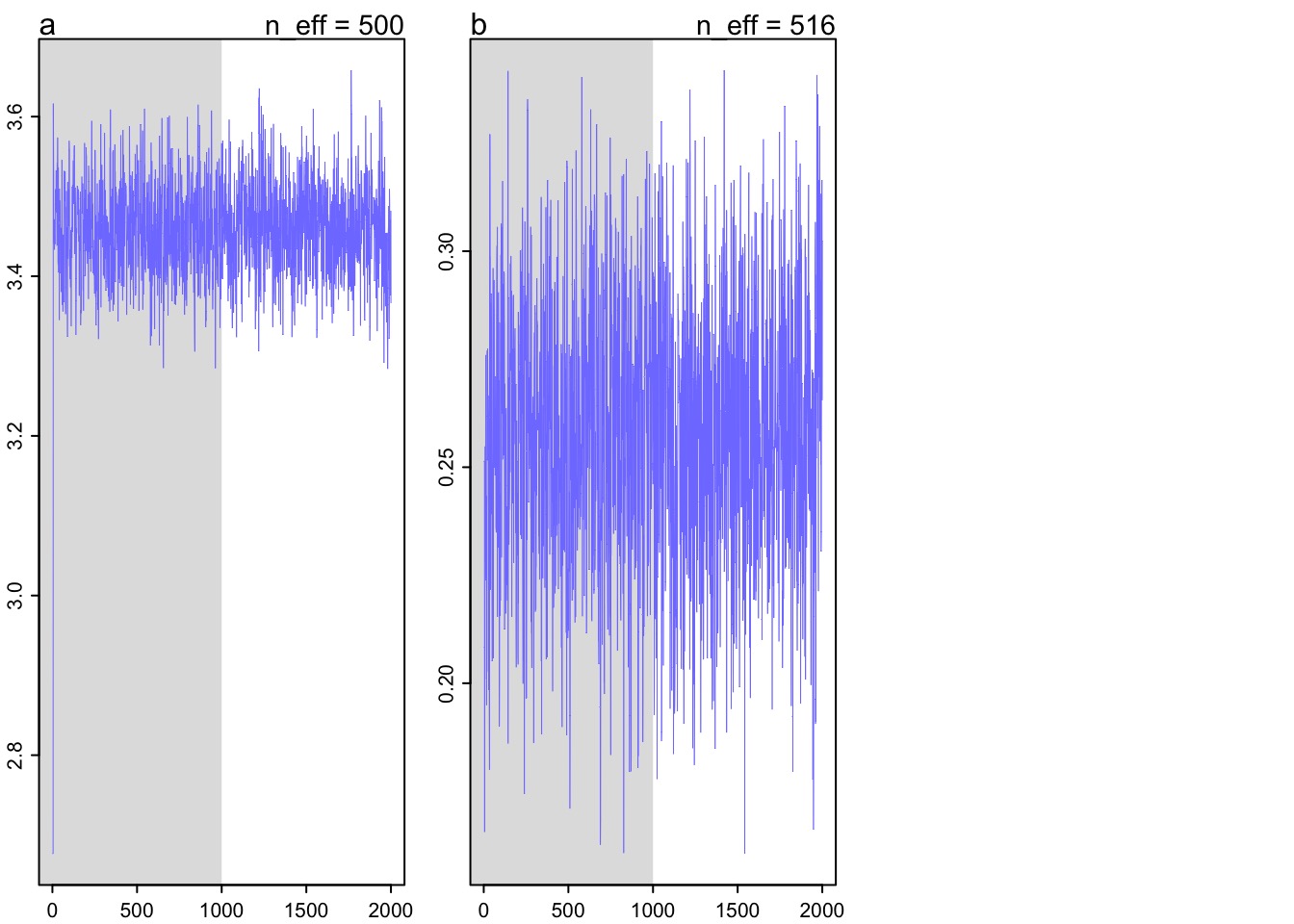
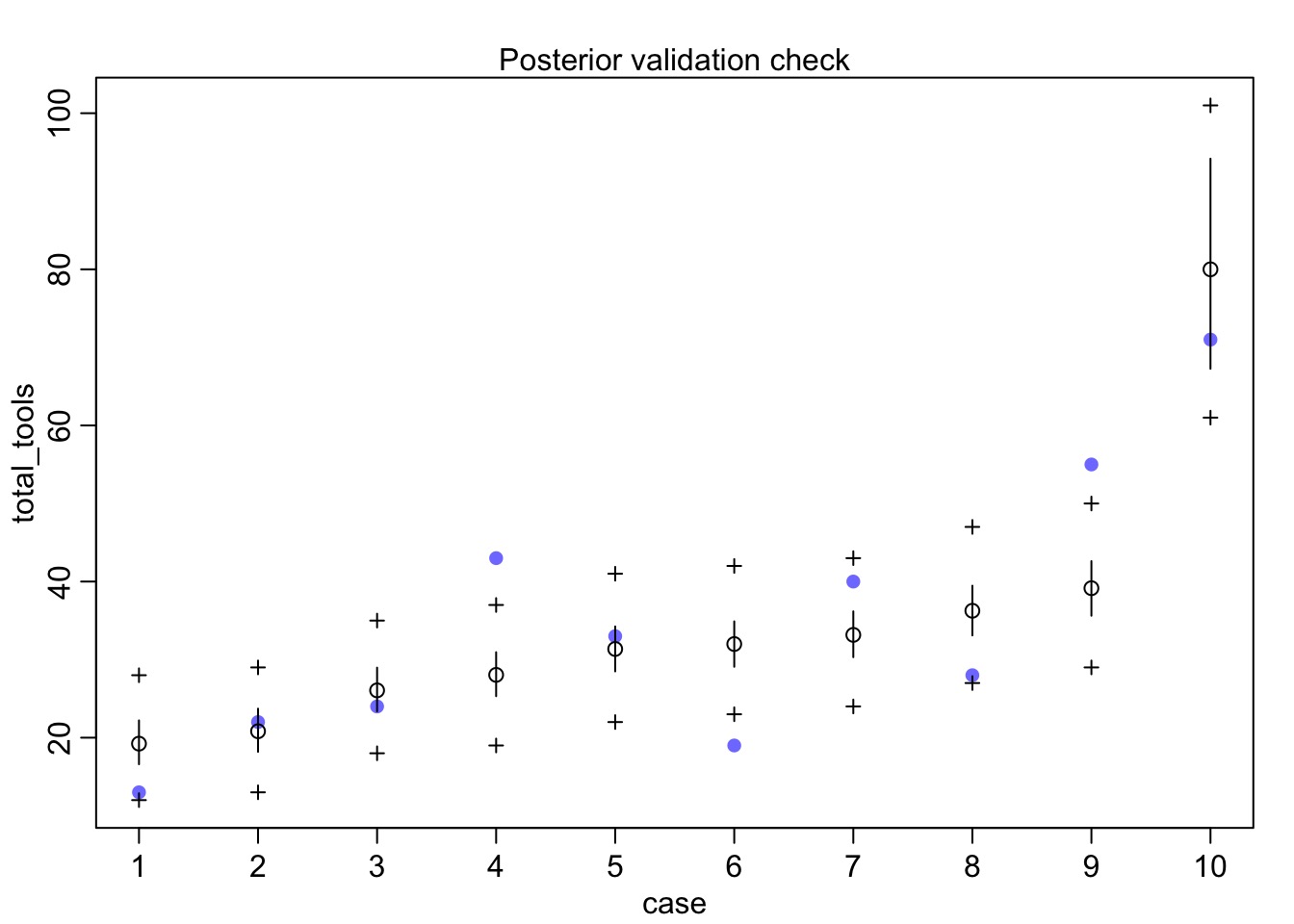
Check your posterior
Check Quantile Residuals
#sim gets predictions
kline_post_pred <- sim(kline_fit, refresh=0)
#figure out how much of each column < observation
quants_kline <- sapply(1:nrow(Kline), function(i){
#what fraction of predictions < the observation
sum(kline_post_pred[,i] < Kline$total_tools[i])/nrow(kline_post_pred)
})
gap::qqunif(quants_kline, logscale=FALSE)Check Quantile Residuals
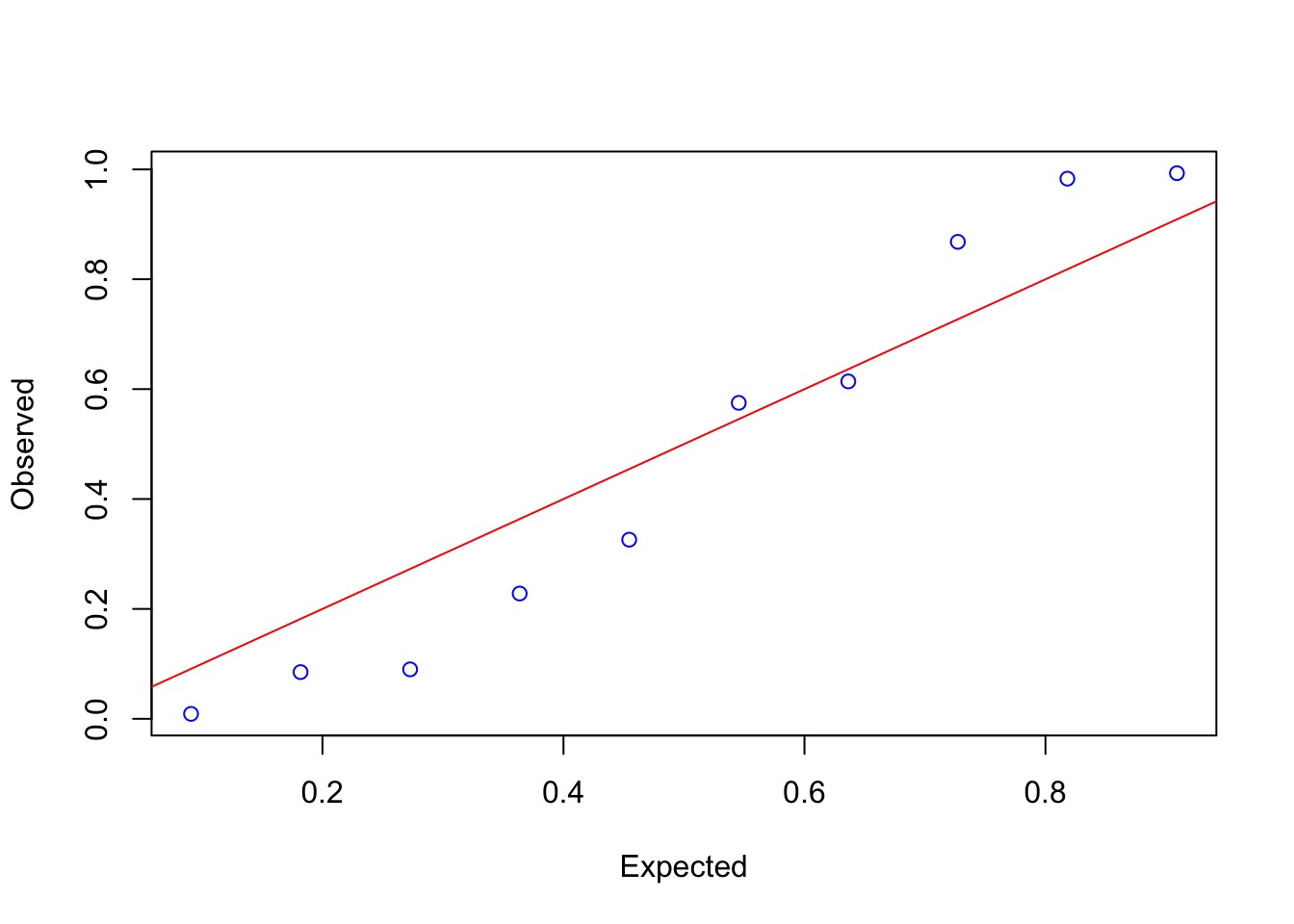
Outcomes?
Mean StdDev lower 0.89 upper 0.89 n_eff Rhat
a 3.46 0.06 3.37 3.56 500 1.00
b 0.26 0.03 0.21 0.31 516 1.01Should the link really be linear?
Kline$P <- log(Kline$population) - mean(log(Kline$population))
kline_linear <- alist(
#likelihood
total_tools ~ dpois(lambda),
#DGP
lambda <- a + b * P,
#priors
a ~ dnorm(3,0.5),
b ~ dnorm(1,0.2)
)
kline_linear_fit <- map2stan(kline_linear, data = Kline)
SAMPLING FOR MODEL 'total_tools ~ dpois(lambda)' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 1.1e-05 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.11 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 0.020341 seconds (Warm-up)
Chain 1: 0.016363 seconds (Sampling)
Chain 1: 0.036704 seconds (Total)
Chain 1:
SAMPLING FOR MODEL 'total_tools ~ dpois(lambda)' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 7e-06 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.07 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: WARNING: No variance estimation is
Chain 1: performed for num_warmup < 20
Chain 1:
Chain 1: Iteration: 1 / 1 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 1e-06 seconds (Warm-up)
Chain 1: 1.9e-05 seconds (Sampling)
Chain 1: 2e-05 seconds (Total)
Chain 1:
[ 100 / 1000 ]
[ 200 / 1000 ]
[ 300 / 1000 ]
[ 400 / 1000 ]
[ 500 / 1000 ]
[ 600 / 1000 ]
[ 700 / 1000 ]
[ 800 / 1000 ]
[ 900 / 1000 ]
[ 1000 / 1000 ]Should the link really be linear?
WAIC pWAIC dWAIC weight SE dSE
kline_fit 85.6 4.3 0 1 9.39 NA
kline_linear_fit 511.6 17.5 426 0 121.30 123.15Your turn - What drives tool use?
culture population contact total_tools mean_TU
1 Malekula 1100 low 13 3.2
2 Tikopia 1500 low 22 4.7
3 Santa Cruz 3600 low 24 4.0
4 Yap 4791 high 43 5.0
5 Lau Fiji 7400 high 33 5.0
6 Trobriand 8000 high 19 4.0