Taking Least Squares From the Ordinary to the General(ized)

Basic Princples of Ordinary Least Squares
- Y is determined by X: p(Y \(|\) X=x)
- The relationship between X and Y is Linear
- The residuals of \(\widehat{Y_i} = \beta_0 + \beta_1 X_i + \epsilon\) are normally distributed
(i.e., \(\epsilon_i \sim\) N(0,\(\sigma\)))
Basic Principles of Ordinary Least Squares
\(\widehat{Y} = \beta_0 + \beta_1 X + \epsilon\) where \(\beta_0\) = intercept, \(\beta_1\) = slope
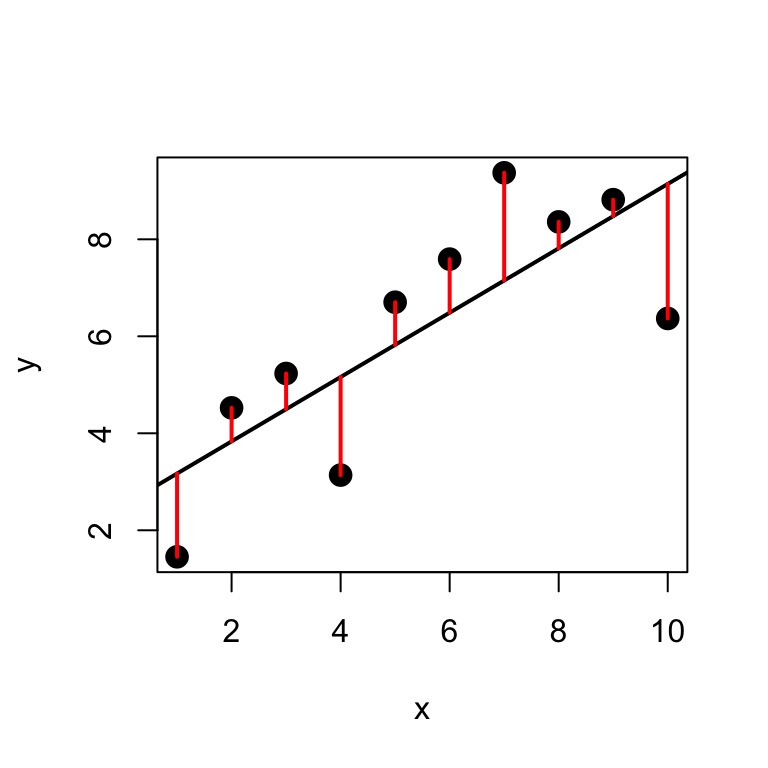
Minimize Residuals defined as \(SS_{residuals} = \sum(Y_{i} - \widehat{Y})^2\)
But…what about this?
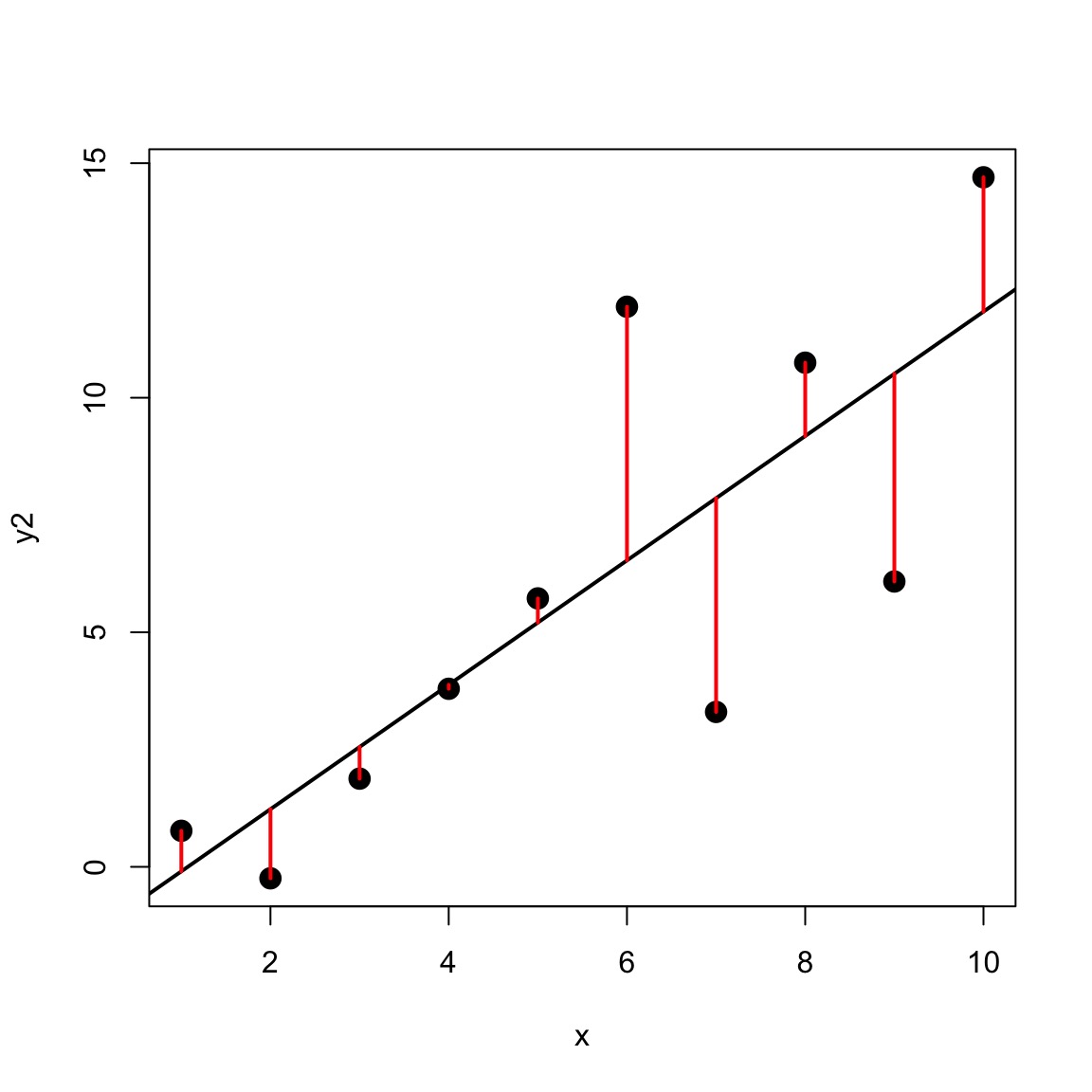
Variance-Mean Relationship
Or…what about this?
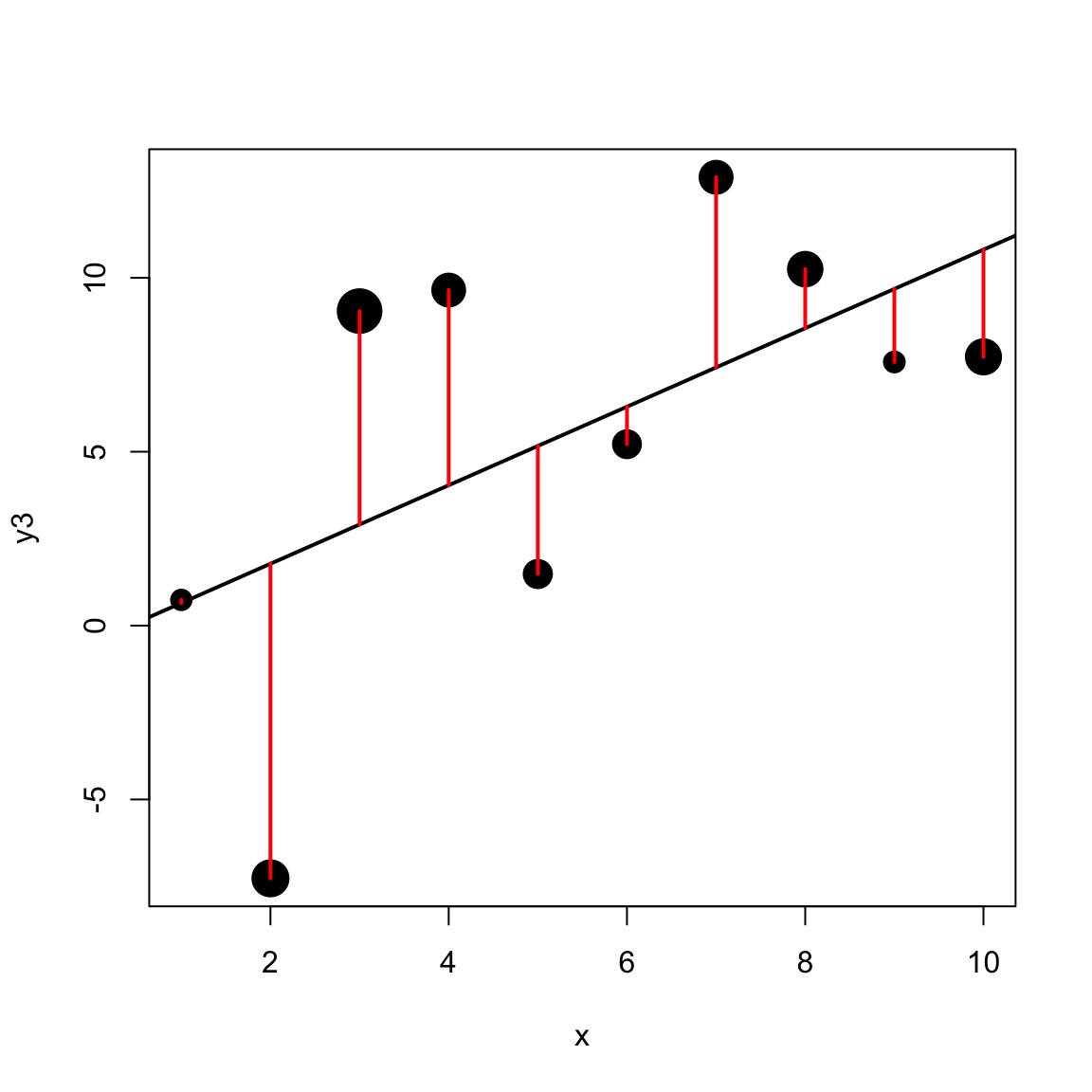
Different precision for different data points
Weighted Least Squares
Why weight?
- Minimized influence of heteroskedasticity
- Increases precision of estimates
- Decreases influence of imprecision in measurements
- Minimize sampling bias
- Other reasons to count some measurements more than others
Weighted Least Squares
\[Y_i = \beta X_i + \epsilon_i\] \[\epsilon_i \sim \mathcal{N}(0, \sigma_i)\]WLS: \(\sigma_i\) is variable
Generalized Least Squares
\[Y_i = \beta X_i + \epsilon_i\] \[\epsilon_i \sim \mathcal{N}(0, \sigma_i)\](\(I_n\) is an identity matrix with n x n dimensions)
In WLS, the diagonal of the \(\sigma\) matrix can be anything
In Generalized Least Squares, the off diagonals can be non-zero, too
How can we use weighting?
- Weighting by variance
- Mean-Variance relationship
- Unequal variance between groups
SQUID!

Somatic v. Reproductive Tissues: Variance
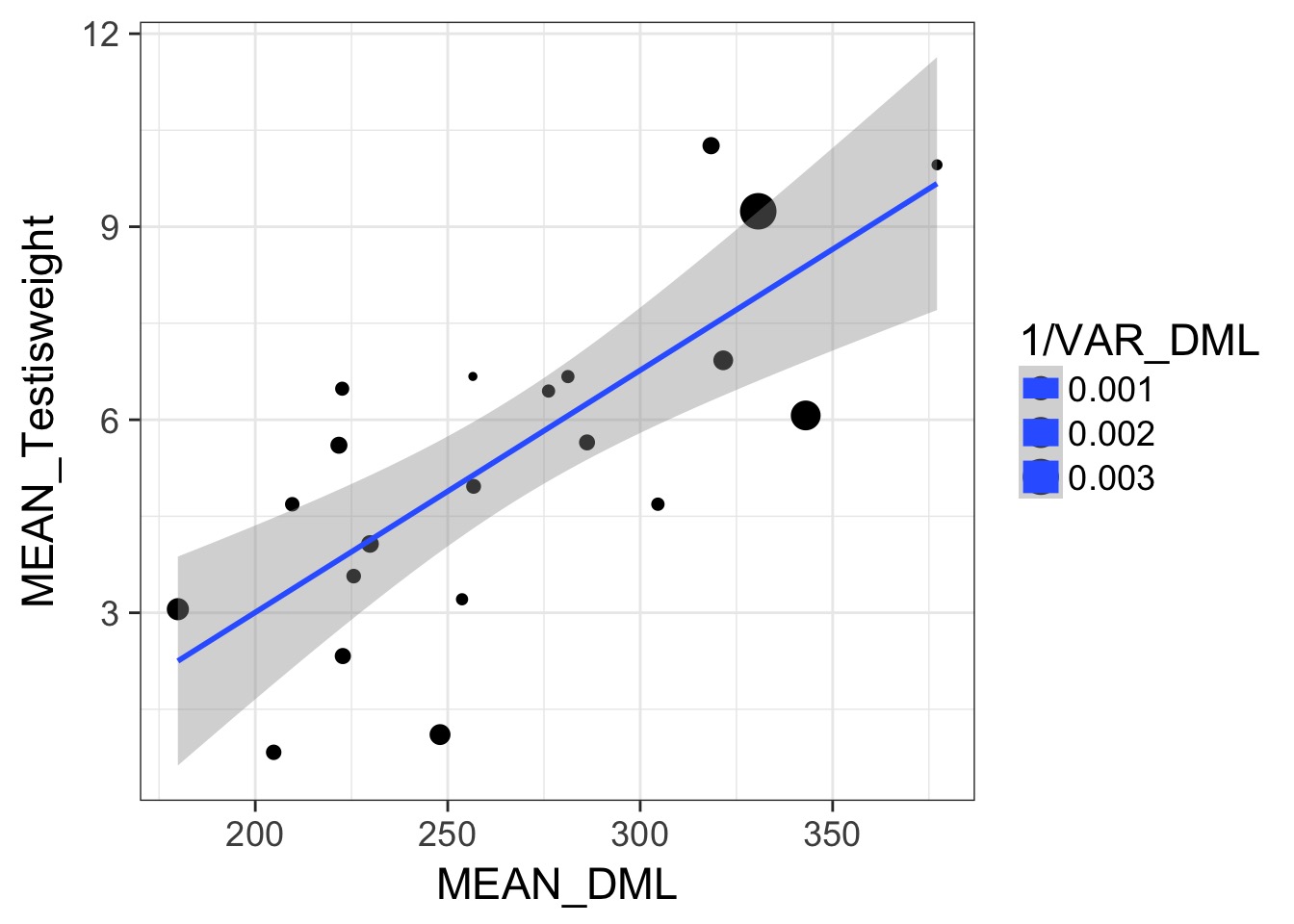
Somatic v. Reproductive Tissues: N
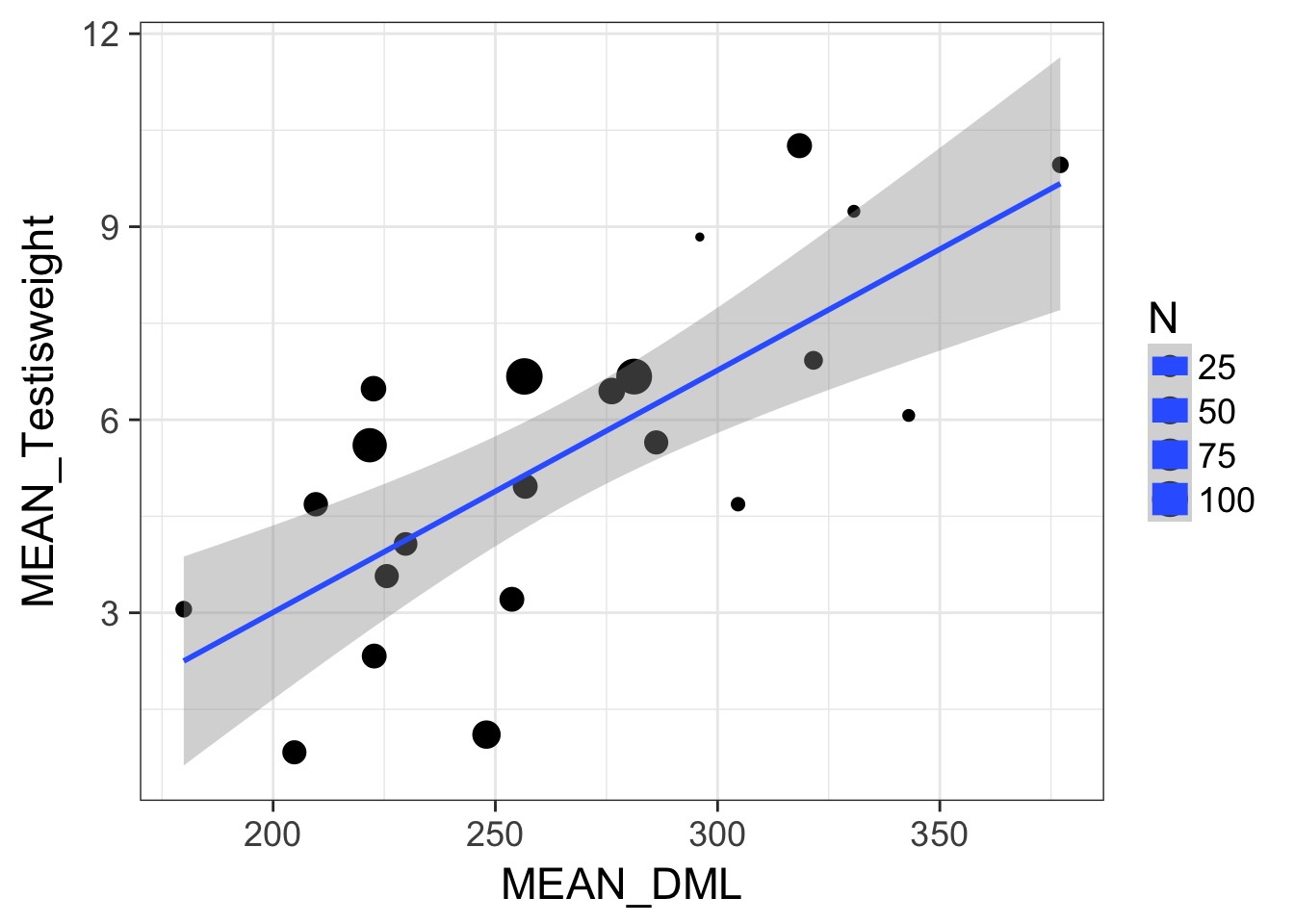
Weighting by Data Quality
- We typically weight by 1/variance (precision)
- Or N, or other estimate of sample precision
- This is different than variance weighted meta-analysis
- We are still estimating a residual error for fits
Implementation
sq_sum <- lm(MEAN_Testisweight ~ MEAN_DML, data=squid_summary,
weights=1/VAR_DML)Did it do anything?
No weights
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -4.5177727 | 2.1081505 | -2.143003 | 0.0445948 |
| MEAN_DML | 0.0376229 | 0.0077701 | 4.842035 | 0.0000989 |
Precision Weighted
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -4.518775 | 2.2041869 | -2.050087 | 0.0544130 |
| MEAN_DML | 0.037140 | 0.0074578 | 4.980010 | 0.0000831 |
Sample Size Weighted
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -5.2808150 | 2.8213670 | -1.871722 | 0.0759446 |
| MEAN_DML | 0.0417104 | 0.0110373 | 3.779050 | 0.0011787 |
How can we use weighting?
- Weighting by variance
- Mean-Variance relationship
- Unequal variance between groups
Looking at Individual Squid
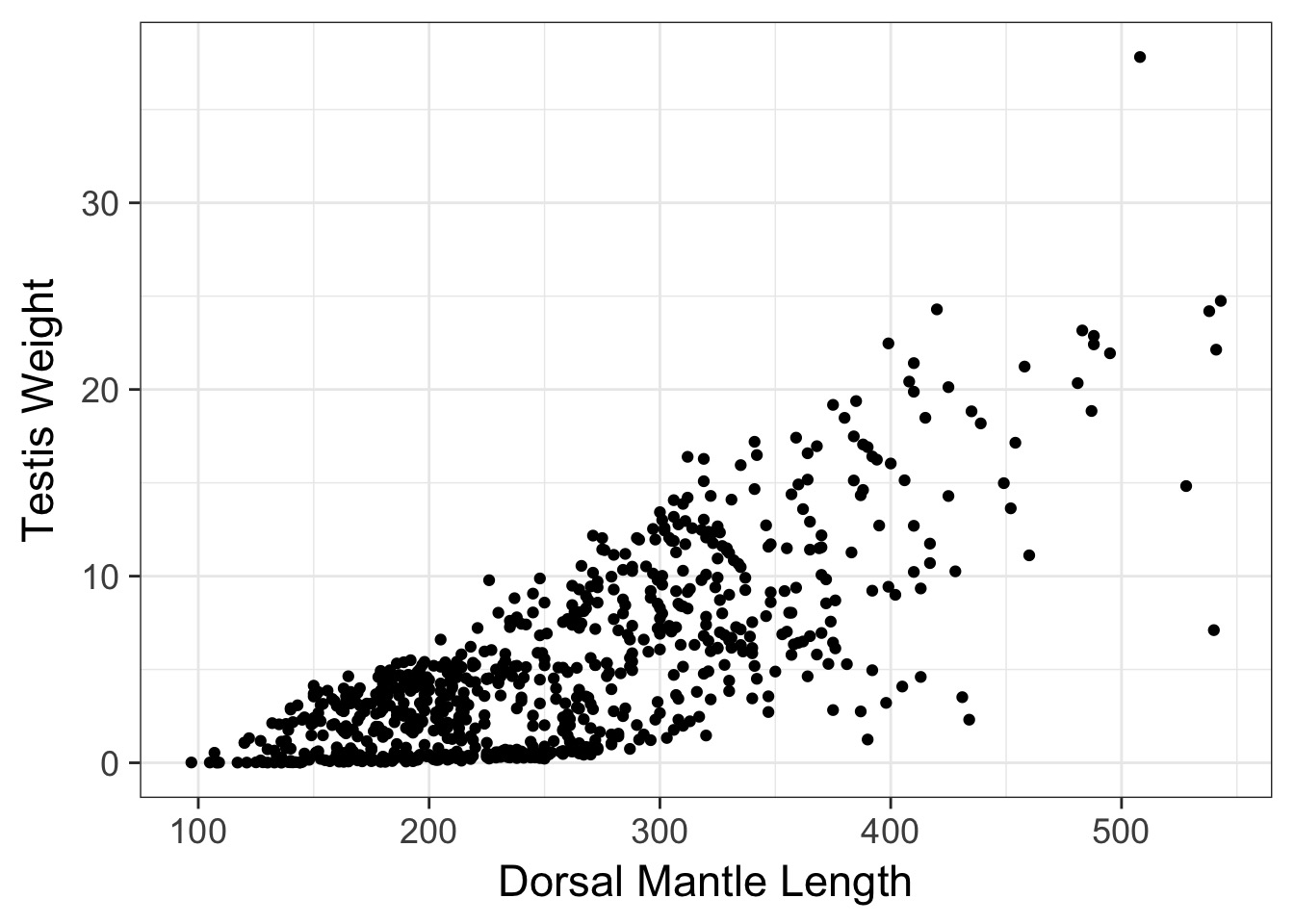
Oh, Yeah, That’s Heteroskedasticity
Oh, Yeah, That’s Heteroskedasticity
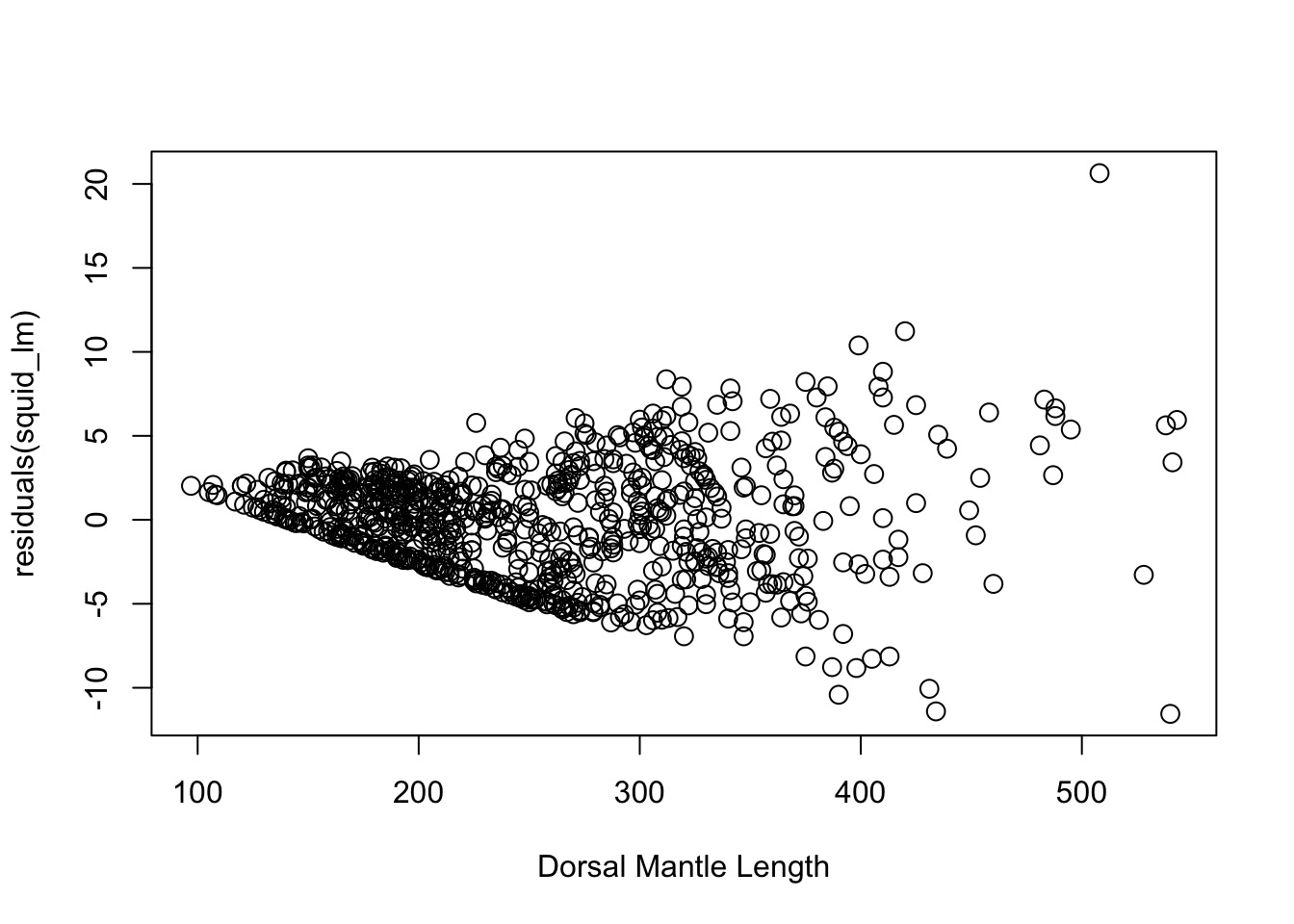
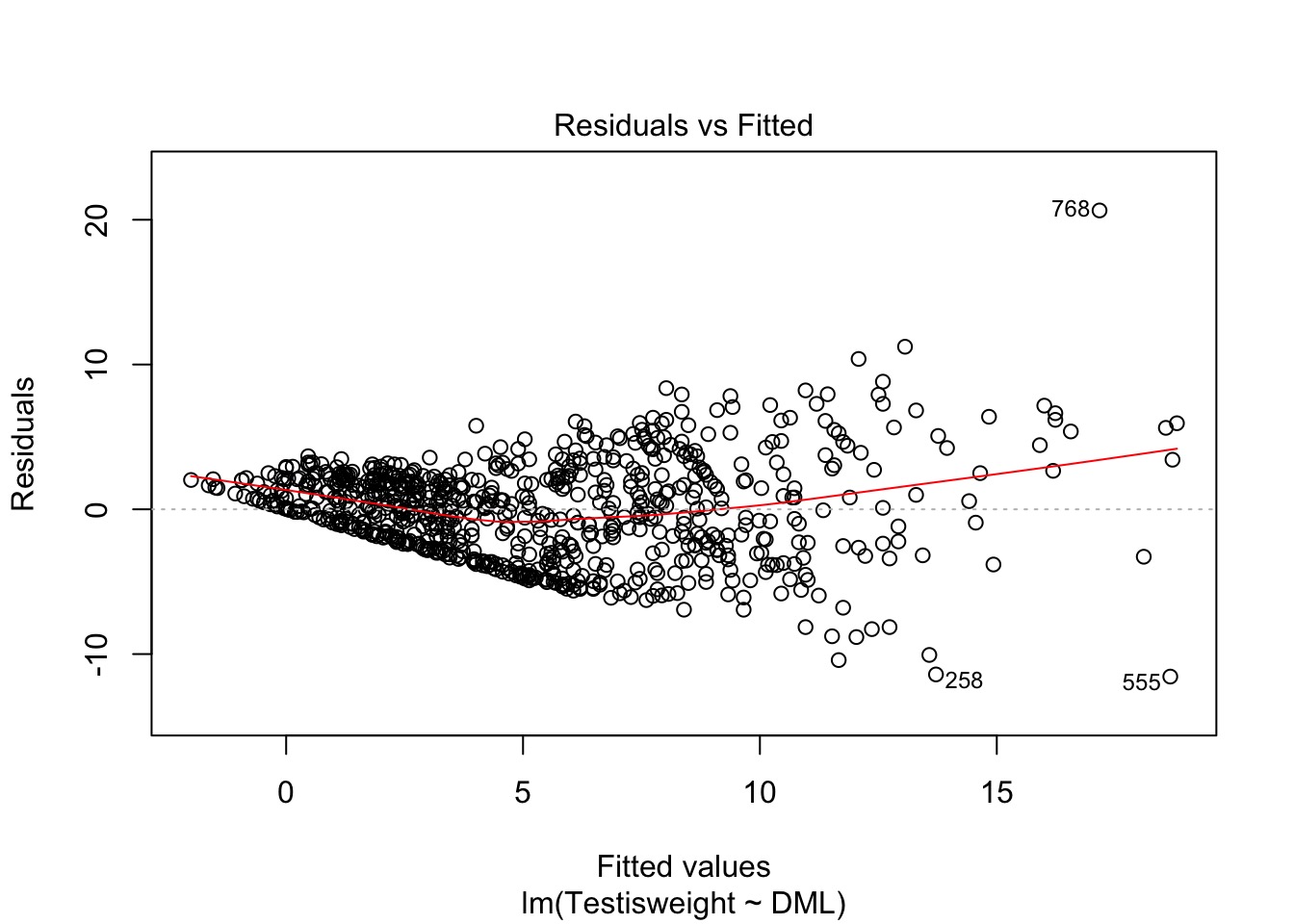
OK, But, We Need a test
- Not every case is clear-cut
- Breusch-Pagan/Godfrey/Koenker test
- Variant of White’s test (you’ll see this)
- Get squared residuals
- Regress on one or more predictors
- Results fail \(\chi^2\) test of heteroskedastic
BP Test
library(lmtest)
squid_lm <- lm(Testisweight ~ DML, data=squid)
bptest(squid_lm)
studentized Breusch-Pagan test
data: squid_lm
BP = 155.31, df = 1, p-value < 2.2e-16Can use BP to Look at Multiple Predictors
squid_lm2 <- lm(Testisweight ~ DML + MONTH, data=squid)
bptest(squid_lm2)
studentized Breusch-Pagan test
data: squid_lm2
BP = 144.22, df = 2, p-value < 2.2e-16Can Look at Contribution of Individual Predictors
bptest(squid_lm2, varformula = ~ DML, data=squid)
studentized Breusch-Pagan test
data: squid_lm2
BP = 140.74, df = 1, p-value < 2.2e-16bptest(squid_lm2, varformula = ~ MONTH, data=squid)
studentized Breusch-Pagan test
data: squid_lm2
BP = 0.086946, df = 1, p-value = 0.7681So How do we Weight This?
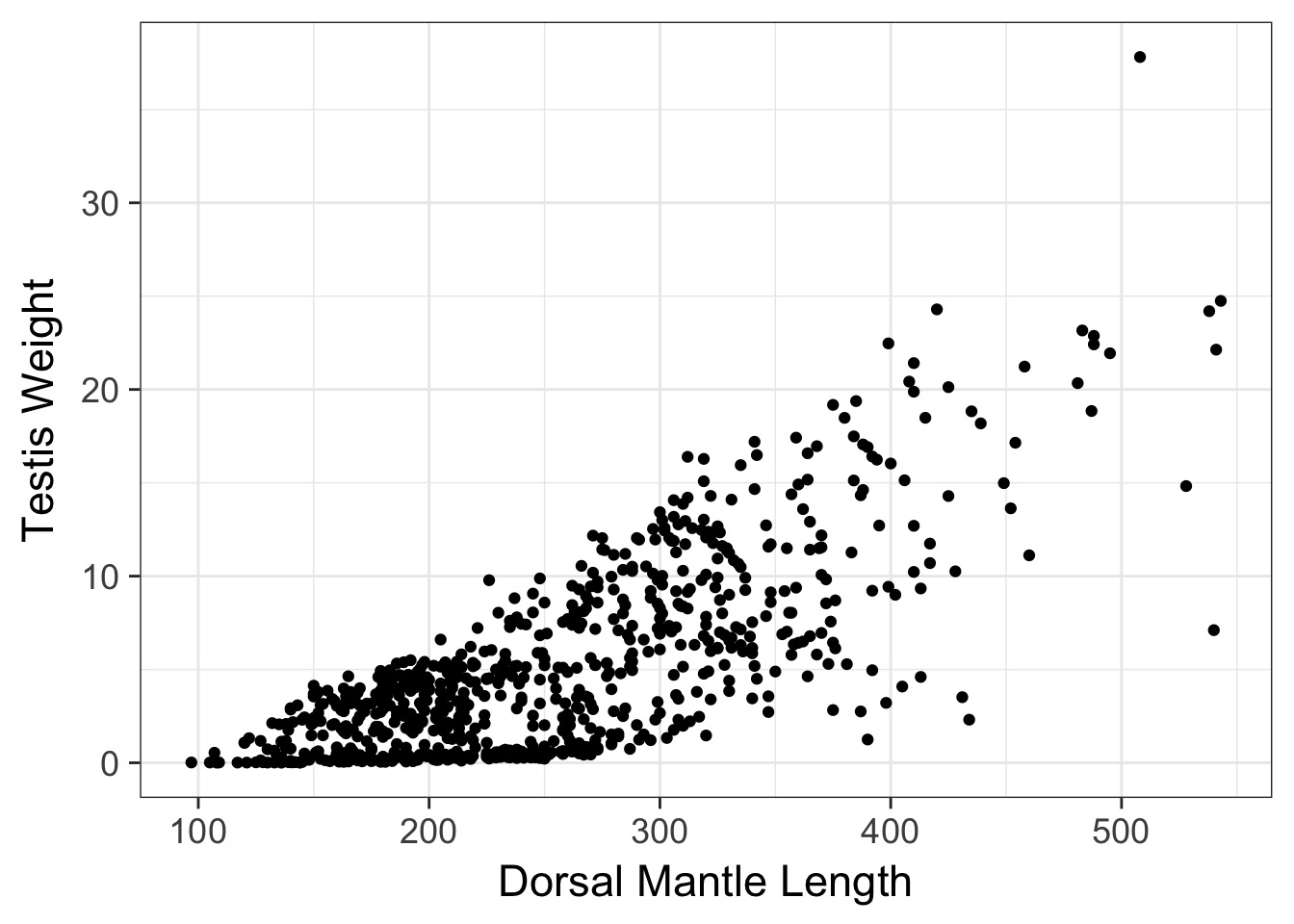
Weighting by a Predictor
- Need to determine the direction variance increases
- Weight by X or 1/X
- Is it a linear or nonlinear relationship between variance and X?
- Is more than one predictor influencing variance (from BP Test/Graphs)
WLS with LM
squid_wls <- lm(Testisweight ~ DML, data=squid,
weights = 1/squid$DML)
Variance increases with DML, so weight decreases
LM is LiMiting
- Need to hand-code the weights
- Diagnostic plots will still look odd
- DHARMa can help
- DHARMa can help
- Cannot easily combine weights from multiple predictors
Enter NLME
- NonLinear Mixed Effects Model package
- We’ll get to what that means
- BUT - also has likelihood-based methods to fit WLS and GLS
- Flexible weighting specification
WLS via GLS in NLME
squid_gls <- gls(Testisweight ~ DML,
data=squid,
weights = varFixed(~ DML))Note: higher value = higher variance - opposite of lm
Compare WLS and GLS
WLS with lm
t test of coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -5.6239370 0.3382932 -16.624 < 2.2e-16 ***
DML 0.0430654 0.0014061 30.627 < 2.2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1GLS with gls
z test of coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -5.6239370 0.3382932 -16.624 < 2.2e-16 ***
DML 0.0430654 0.0014061 30.627 < 2.2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Different Variance Structures
VarFixed- Linear continuous varianceVarPower- Variance increases by a powerVarExp- Variance exponentiatedVarConstPower- Variance is constant + powerVarIdent- Variance differs by groupsVarComb- Combines different variance functions
Multiple Sources of Heteroskedasticity
Let’s add month! If it was another driver…
squid_gls_month <- gls(Testisweight ~ DML, data=squid,
weights = varComb(varFixed(~DML),
varFixed(~MONTH)))Weighted v. Naieve Fit
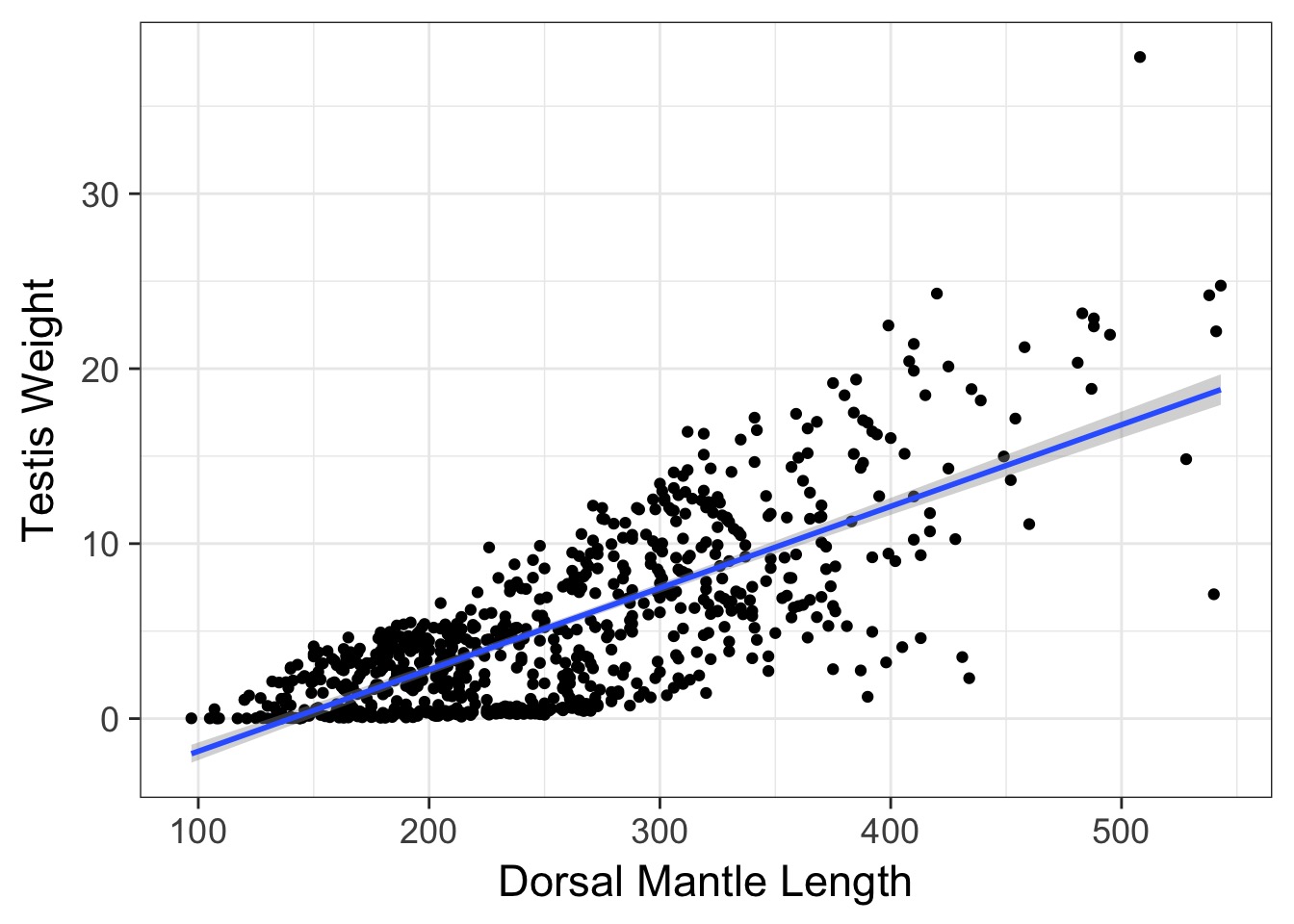
Weighted v. Naieve Fit
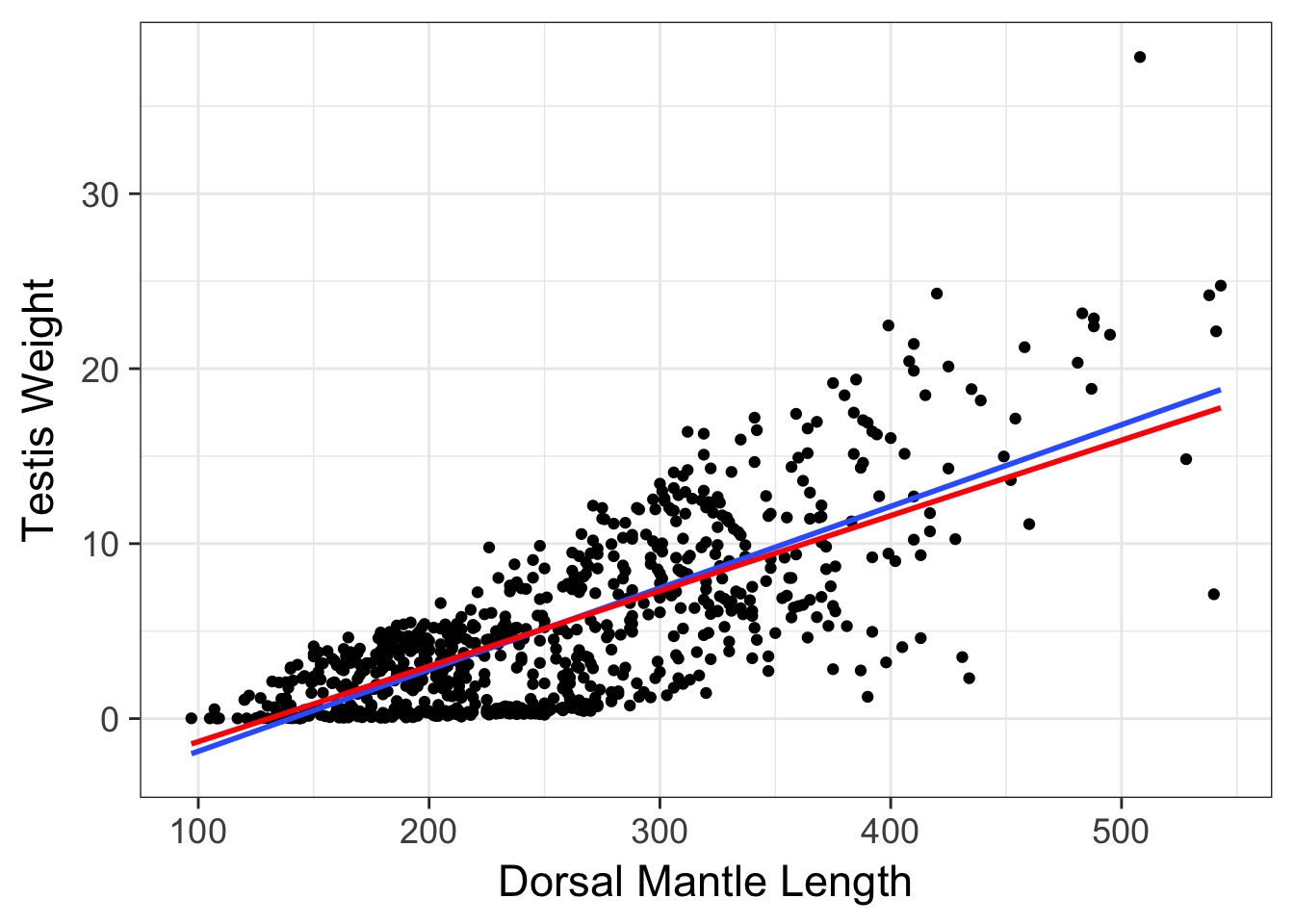
How can we use weighting?
- Weighting by variance
- Mean-Variance relationship
- Unequal variance between groups
Testis Weight by Month
squid <- mutate(squid, fMONTH = factor(MONTH))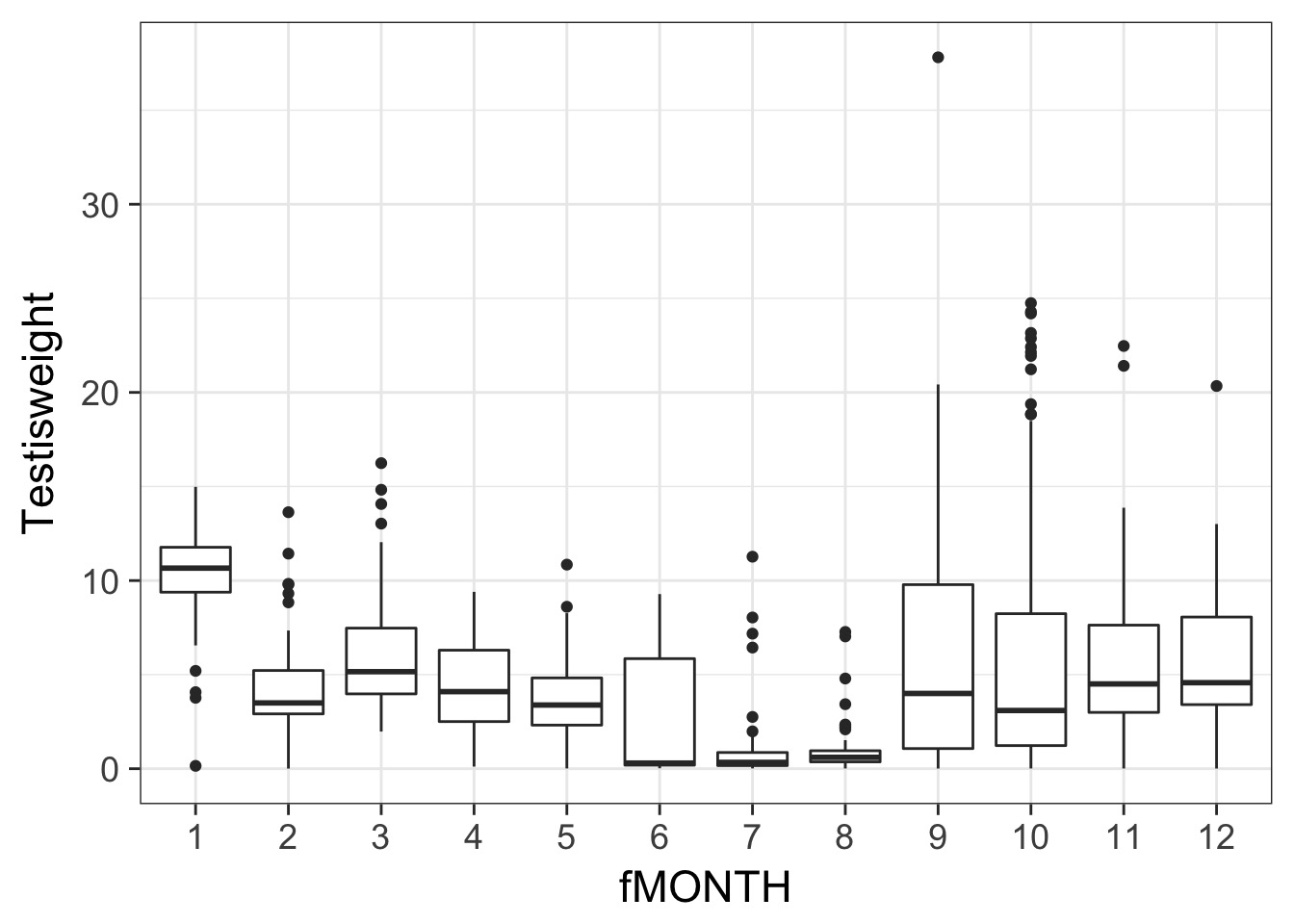
A Classic ANOVA problem
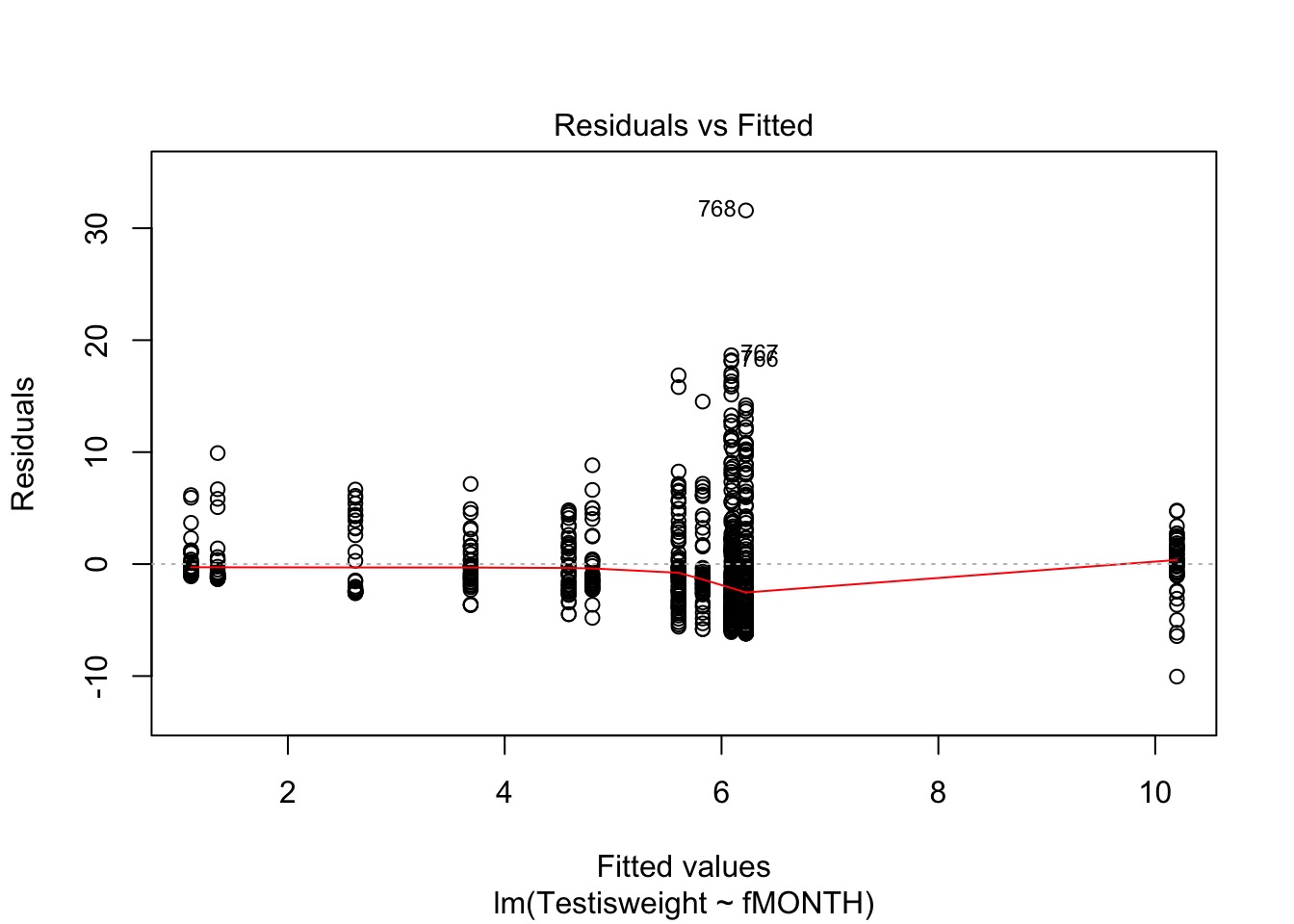
Wowsers - Unequal Variances
| MONTH | var_weight |
|---|---|
| 1 | 8.216093 |
| 2 | 9.491376 |
| 3 | 9.456260 |
| 4 | 6.663934 |
| 5 | 5.513464 |
| 6 | 10.884092 |
| 7 | 6.635753 |
| 8 | 2.274724 |
| 9 | 41.502657 |
| 10 | 46.016877 |
| 11 | 17.488115 |
| 12 | 17.734229 |
Is this a probem?
bptest(squid_month)
studentized Breusch-Pagan test
data: squid_month
BP = 68.375, df = 11, p-value = 2.485e-10Solution: Weight by Month
vMonth <- varIdent(form = ~ 1 | fMONTH)- Note
1 | xform, different variance for different strata
Fit and Estimate SD
squid_month_gls <- gls(Testisweight ~ fMONTH,
data=squid,
weights=vMonth)Summary of results
Generalized least squares fit by REML
Model: Testisweight ~ fMONTH
Data: squid
AIC BIC logLik
4307.961 4419.034 -2129.98
Variance function:
Structure: Different standard deviations per stratum
Formula: ~1 | fMONTH
Parameter estimates:
2 9 12 11 8 10 5
1.0000000 2.0910010 1.3669006 1.3573650 0.4895507 2.2018372 0.7621603
7 6 4 1 3
0.8361282 1.0708259 0.8379144 0.9303731 0.9981328
Coefficients:
Value Std.Error t-value p-value
(Intercept) 10.200311 0.4272917 23.872011 0
fMONTH2 -5.391282 0.6795202 -7.933953 0
fMONTH3 -4.094684 0.5555744 -7.370183 0
fMONTH4 -5.609615 0.5722338 -9.803013 0
fMONTH5 -6.514285 0.5724285 -11.380085 0
fMONTH6 -7.577601 0.6848334 -11.064882 0
fMONTH7 -8.847744 0.6016011 -14.706993 0
fMONTH8 -9.093561 0.4757355 -19.114743 0
fMONTH9 -3.975147 0.7016302 -5.665587 0
fMONTH10 -4.109998 0.7252503 -5.667006 0
fMONTH11 -4.596277 0.6174994 -7.443371 0
fMONTH12 -4.373843 0.7482711 -5.845265 0
Correlation:
(Intr) fMONTH2 fMONTH3 fMONTH4 fMONTH5 fMONTH6 fMONTH7 fMONTH8
fMONTH2 -0.629
fMONTH3 -0.769 0.484
fMONTH4 -0.747 0.470 0.574
fMONTH5 -0.746 0.469 0.574 0.557
fMONTH6 -0.624 0.392 0.480 0.466 0.466
fMONTH7 -0.710 0.447 0.546 0.530 0.530 0.443
fMONTH8 -0.898 0.565 0.691 0.671 0.670 0.560 0.638
fMONTH9 -0.609 0.383 0.468 0.455 0.455 0.380 0.433 0.547
fMONTH10 -0.589 0.370 0.453 0.440 0.440 0.368 0.418 0.529
fMONTH11 -0.692 0.435 0.532 0.517 0.517 0.432 0.491 0.622
fMONTH12 -0.571 0.359 0.439 0.426 0.426 0.356 0.406 0.513
fMONTH9 fMONTH10 fMONTH11
fMONTH2
fMONTH3
fMONTH4
fMONTH5
fMONTH6
fMONTH7
fMONTH8
fMONTH9
fMONTH10 0.359
fMONTH11 0.421 0.408
fMONTH12 0.348 0.336 0.395
Standardized residuals:
Min Q1 Med Q3 Max
-3.5056004 -0.6728254 -0.3252821 0.4762647 4.9030307
Residual standard error: 3.080871
Degrees of freedom: 768 total; 756 residualCompare with Unweighted Fit
squid_lm <- gls(Testisweight ~ fMONTH, data=squid)
anova(squid_lm, squid_month_gls) Model df AIC BIC logLik Test L.Ratio
squid_lm 1 13 4555.853 4616.017 -2264.926
squid_month_gls 2 24 4307.961 4419.034 -2129.980 1 vs 2 269.892
p-value
squid_lm
squid_month_gls <.0001A Clammy Example
- Clams.txt has data on the length-AFDM relationship of Wedge clames in Argentina each month.
- Useread.delimto read it in
- Evaluate the length-biomass relationship
- Test for Heteroskedasticity
- Correct for it
- Do the same for just month
- You’ll have to make it a factor
- Combine them into one model!
- usevarComb